
Ampullary and pancreatic adenocarcinoma—a comparative study
Introduction
Pancreatic ductal adenocarcinoma (PDAC) and ampullary adenocarcinoma (AAC) are 2 gastrointestinal cancers that share overlapping symptoms (1). Although some studies have proposed the hypothesis of their differences in pathogenesis, prognosis and molecular profile; they remain treated similarly by pancreaticoduodenectomy followed by adjuvant chemotherapy (2,3).
Epidemiologically, PDAC was estimated to be the 7th reason of mortality by cancer worldwide in 2014 (4). Projection studies show that it could become the 2nd leading cause of cancer deaths in 2020 and that its incidence is increasing because of the transfer of risk factors like poor eating habits, sedentary lifestyle, obesity, and smoking from the developing to the less developing countries (5). Only few studies described AAC characteristics. These were assimilated, in the majority of cases, to PDAC; although it has a better prognosis than PDAC. In fact, 5-year survival rate of AAC is wide-ranging (from 36.8% to 75.2%). In addition, the genetic characteristics of these 2 tumors remain unclear and ambiguous (1,6,7).
Along these lines, the aim of this study was to participate to the consolidation of AAC biology understanding using a comparative approach of clinicopathological parameters of 39 cases of PDAC and 21 cases of AAC reclassified previously using immunohistochemistry (IHC); additionally to analysis of mutational status of 3 oncogenes (KRAS, NRAS and BRAF).
Methods
This retrospective study was approved by the Ethics Committee of Habib Thameur Hospital in Tunis (HTHEC). Formalin-fixed paraffin-embedded (FFPE) specimens from 39 PDAC and 21 AAC, resected from 2000 to 2016, were obtained from the archival block of Pathology Department. All specimens were fixed in 10% buffered formalin. The cases were reviewed separately by two experimented pathologists (R Jouini, E BenBrahim) based on an evaluation of hematoxylin-eosin (H&E) stains. Clinical and epidemiological parameters (age, sex, tumor size, tumor localization, TNM stage, differentiation, vascular emboli and perineural invasion) were determined from patients’ reports. AAC reclassification was based on a confrontation of H&E and IHC evaluation as described in our previous study (8). Genetic analysis of KRAS, NRAS and BRAF included mutational status investigation of 9 hotspot mutation sites covering codons 12-13, codons 59-61, codon 117 and codon 146 of both KRAS and NRAS; as well as codon 600 of BRAF; was performed using PCR, gel electrophoresis and pyrosequencing via a PyroMark Q24 instrument as detailed in our previous study (9). SPSS 20.0 (SPSS, Inc., USA) was used for comparison with a significative P value less than 0.05.
Results
PDAC and AAC patients’ characteristics and genetic analysis results are detailed and discussed in our previous studies (8,9). Briefly, AAC cases were reclassified to 15 (71.5%) pancreatobiliary (PB), 2 (9.5%) intestinal (IT) and 4 (19%) mixed (M). Table 1 shows mutational status and mutation classes’ distribution among PDAC, AAC and PB subtype.
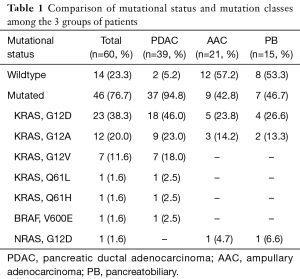
Full table
In the comparative study, tumor site (PDAC vs. AAC) was significantly associated to tumor differentiation (P<0.001), perineural invasion (P<0.001), vascular emboli (P=0.001), T stage (P=0.007), N stage (P=0.001) and mutational status (P<0.001) (Table 2).
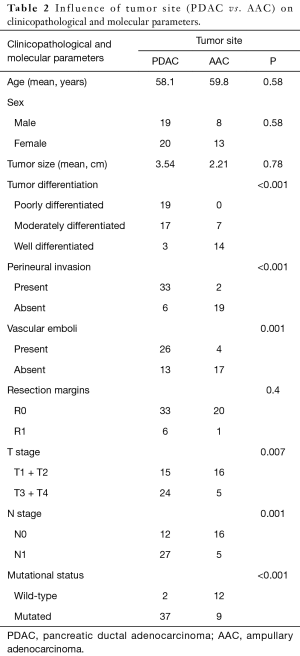
Full table
When the cases belonging to IT and M subtypes were discarded, influence of tumor type on clinicopathological and molecular parameters of patients hadn’t changed considerably. In fact, tumor type (PDAC vs. PB subtype) was significantly associated to tumor size (P=0.001), tumor differentiation (P<0.001), perineural invasion (P<0.001), vascular emboli (P=0.001), T stage (P=0.033), N stage (P<0.001) and mutational status (P<0.001) (Table 3).
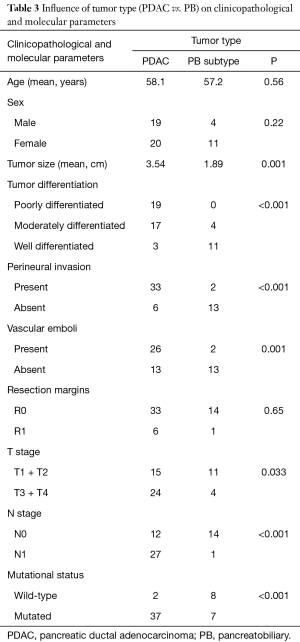
Full table
Discussion
Attempts to classify AAC face several challenges. Moreover, genetic characteristics of different subtypes remain unclear and ambiguous (1,7). By analysing KRAS, NRAS and BRAF status in AAC and PDAC, we hoped to participate in the consolidation of AAC pathogenesis comprehension.
The majority (94.8%) of our PDAC harbored KRAS mutations dominated by codon 12 mutation and G12D was the most predominant mutational class as described in literature data (10-13). Concerning AAC, their epidemiological characteristics are not well known because of its rarity. To our knowledge, no epidemiological study in Tunisia has been published describing its features. Our cohort comprised 62% of women; unlike the majority of literature data where men were still the most affected by this class of tumors (14-16). A study of 256 French patients didn’t report any survival improvement in over 34 years. Tumor stage, nodal invasion status were classified as important prognostic factors (17). In more recent studies, patients survival was associated with TNM stage, perineural invasion, vascular emboli, tumor differentiation, pancreatic invasion and tumor size (18-22).
After confrontation of IHC and H&E evaluation, the majority of our cases (72%) were reclassified as PB; in line with previous studies which found very wide-ranging frequencies of PB subtype: from 22% to 72% (3,7,18-34). Three factors could explain this range: first, the rarity of this cancer and the small number of patients in most of studies; second, the heterogeneity of this pathology that divides this small population and makes studying a specific subtype very difficult; third, the absence of a common classification tools. In fact, IHC markers vary from one study to another with use of MUC1, MUC2, MUC4, MUC5CA, CDX2, CK7 and CK20, etc. Also, we can highlight the ambiguous definition of M subtype. In some studies, 6% to 9% of the 2 epithelia are required for the diagnosis of this subtype while others require a minimum of 10% even 25% (22).
Using a whole-exome-sequencing (WES) of 60 AAC, a study identified 24 recurrent mutated genes. Of those, 9 were significantly mutated in the PB subtype including KRAS (29). These data confirmed a previous study suggesting that incidence of mutations frequently implicated in PDAC carcinogenesis, particularly KRAS, correlate with PB subtype, although they are not specific for it (24). In our AAC series, KRAS was mutated in 47% of PB subtype. Literature data confirm that KRAS is mutated in 20% to 61% in PB subtype (19,22,25,30,35).
Comparing PDAC and AAC; we found a significant correlation between tumor site and tumor differentiation, perineural invasion, vascular emboli, T stage, N stage and mutational status. When cases belonging to IT and M subtypes were discarded; these differences changed only slightly. Thereby, tumor type (PDAC vs. PB) was significantly correlated with tumor differentiation, perineural invasion, vascular emboli, T stage, N stage, and mutational status, as well as tumor size. Indeed, this latter was almost double in PDAC group (3.45 cm) compared to AAC group (1.89 cm) with a significant P value (0.001). This difference could be explained by the early obstruction of bile duct or pancreatic duct. With these results, we do not share the proposal to continue a common treatment for PB and PDAC like suggested by other studies (18,20-22,25-27,32,33,36). Moreover, significant difference in survival between AAC and PDAC was found (3).
On the other hand, lack of common anatomical definition between authors presents another important challenge in AAC studies. In fact, some authors defined PDAC and AAC like two entities belonging to periampullary cancer (1-3,23,35). Other groups exclude PDAC of pancreas head from this group (16,36,37).
Conclusions
We think that we should differentiate between PDAC and AAC since even in comparison with PB subtype alone, PDAC remains worse in terms of clinicopathological and molecular characteristics. Small number of our cohort preclude consistent conclusions. Further studies are needed to better understand AAC biology.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by Habib Thameur Hospital ethics committee (HTHEC, No. HTHEC-2017-03). As this was a retrospective study, informed consent is not required.
References
- Uomo G. Periampullary carcinoma: some important news in histopathology. JOP 2014;15:213-5. [PubMed]
- Baghmar S, Agrawal N, Kumar G, et al. Prognostic factors and the role of adjuvant treatment in periampullary carcinoma: a single-centre experience of 95 Patients. J Gastrointest Cancer 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Sánchez-García J, Candanedo-González F, Félix-Félix AK, et al. Retrospective cohort of pancreatic and Vater ampullary adenocarcinoma from a reference center in Mexico. Ann Med Surg (Lond) 2018;30:7-12. [Crossref] [PubMed]
- Bernard SW, Christopher PW. World cancer report. IARC WHO 2014.
- Ferlay J, Partensky C. Séance thématique: Progrès dans la prise en charge des adénocarcinomes du pancréas. L’augmentation inquiétante de l’incidence et de la mortalité du cancer du pancréas à l’échelle mondiale. Bull Acad Natl Med 2017;201:1-8.
- Junrungsee S, Kittivarakul E, Ko-iam W, et al. Prognostic Factors and Survival of Patients with Carcinoma of the Ampulla of Vater after Pancreaticoduodenectomy. Asian Pac J Cancer Prev 2017;18:225-9. [PubMed]
- Perysinakis I, Margaris I, Kouraklis G. Ampullary cancer--a separate clinical entity? Histopathology 2014;64:759-68. [Crossref] [PubMed]
- Ferchichi M, Jouini R, Ayari I, et al. KRAS, NRAS and BRAF analysis of ampullary adenocarcinoma classified using CK7, CK20, MUC1 and MUC2. J Gastrointest Oncol 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Ferchichi M, Jouini R, Ayari I, et al. Kras, epidermal growth factor receptor, and p53, but not Nras or Braf, biomarkers are frequently altered in pancreatic adenocarcinoma and precursor lesions. Drug Invent 2017;9:88-96.
- Bryant KL, Mancias JD, Kimmelman AC, et al. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci 2014;39:91-100. [Crossref] [PubMed]
- Saiki Y, Horii A. Molecular pathology of pancreatic cancer. Pathol Int 2014;64:10-9. [PubMed]
- Schultz NA, Roslind A, Christensen J, et al. Frequencies and prognostic role of KRAS and BRAF mutations in patients with localized pancreatic and ampullary adenocarcinomas. Pancreas 2012;41:759-66. [PubMed]
- Azuara D, Ginesta MM, Gausachs M, et al. Nanofluidic digital PCR for KRAS mutation detection and quantification in gastrointestinal cancer. Clin Chem 2012;58:1332-41. [Crossref] [PubMed]
- Gnoni A, Licchetta A, Scarpa A, et al. Carcinogenesis of pancreatic adenocarcinoma: precursor lesions. Int J Mol Sci 2013;14:19731-62. [Crossref] [PubMed]
- Benhamiche AM, Jouve JL, Manfredi S, et al. Cancer of the ampulla of Vater: results of a 20-year population-based study. Eur J Gastroenterol Hepatol 2000;12:75-9. [Crossref] [PubMed]
- Malla BR, Rodrigues G. Periampullary carcinoma: an audit of an institutional experience with literature review. Surgical Chronicles 2017;22:4-6.
- Rostain F, Hamza S, Drouillard A, et al. Trends in incidence and management of cancer of the ampulla of Vater. World J Gastroenterol 2014;20:10144-50. [Crossref] [PubMed]
- Morini S, Perrone G, Borzomati D, et al. Carcinoma of the ampulla of Vater: morphological and immunophenotypical classification predicts overall survival. Pancreas 2013;42:60-6. [Crossref] [PubMed]
- Mafficini A, Amato E, Cataldo I, et al. Ampulla of vater carcinoma: sequencing analysis identifies TP53 status as a novel independent prognostic factor and potentially actionable ERBB, PI3K, and WNT pathways gene mutations. Ann Surg 2018;267:149-56. [Crossref] [PubMed]
- Valsangkar NP, Ingkakul T, Correa-Gallego C, et al. Survival in ampullary cancer: potential role of different KRAS mutations. Surgery 2015;157:260-8. [Crossref] [PubMed]
- Schueneman A, Goggins M, Ensor J, et al. Validation of histomolecular classification utilizing histological subtype, MUC1 and CDX2 for prognostication of resected ampullary adenocarcinoma. Br J Cancer 2015;113:64-8. [Crossref] [PubMed]
- Asano E, Okano K, Oshima M, et al. Phenotypic characterization and clinical outcome in ampullary adenocarcinoma. J Surg Oncol 2016;114:119-27. [Crossref] [PubMed]
- Chandrasegaram MD, Chen JW, Price TJ, et al. Advances in molecular pathology and treatment of periampullary cancers. Pancreas 2016;45:32-9. [Crossref] [PubMed]
- Hechtman JF, Liu W, Sadowska J, et al. Sequencing of 279 cancer genes in ampullary carcinoma reveals trends relating to histologic subtypes and frequent amplification and overexpression of ERBB2 (HER2). Mod Pathol 2015;28:1123-29. [Crossref] [PubMed]
- Mikhitarian K, Pollen M, Zhao Z, et al. Epidermal growth factor receptor signaling pathway is frequently altered in ampullary carcinoma at protein and genetic levels. Mod Pathol 2014;27:665-74. [Crossref] [PubMed]
- Chang DK, Jamieson NB, Johns AL, et al. Histomolecular phenotypes and outcome in adenocarcinoma of the ampulla of Vater. J Clin Oncol 2013;31:1348-56. [Crossref] [PubMed]
- Perysinakis I, Minaidou E, Leontara V, et al. Differential expression of β-catenin, EGFR, CK7, CK20, MUC1, MUC2, and CDX2 in intestinal and pancreatobiliary- type ampullary carcinomas. Int J Surg Pathol 2017;25:31-40. [Crossref] [PubMed]
- Kawabata Y, Tanaka T, Nishisaka T, et al. Cytokeratin 20 (CK20) and apomucin 1 (MUC1) expression in ampullary carcinoma: correlation with tumor progression and prognosis. Diagn Pathol 2010;5:75. [Crossref] [PubMed]
- Tan J, Tan P, Tean Teh B. Defining the molecular alterations of ampullary carcinoma. Cancer Cell 2016;29:135-6. [Crossref] [PubMed]
- Kohler I, Jacob D, Budzies J, et al. Phenotypic and genotypic characterization of carcinomas of the papilla of Vater has prognostic and putative therapeutic implications. Am J Clin Pathol 2011;135:202-11. [Crossref] [PubMed]
- Schultz NA, Werner J, Willenbrock H, et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol 2012;25:1609-22. [Crossref] [PubMed]
- Leo JM, Kalloger SE, Peixoto RD, et al. Immunophenotyping of ampullary carcinomata allows for stratification of treatment specific subgroups. J Clin Pathol 2016;69:431-9. [Crossref] [PubMed]
- Overman MJ, Zhang J, Kopetz S, et al. Gene expression profiling of ampullary carcinomas classifies ampullary carcinomas into biliary-like and intestinal-like subtypes that are prognostic of outcome. PLoS One 2013;8:1-10. [Crossref] [PubMed]
- Lau SK, Weiss LM, Chu PG. Differential expression of MUC1, MUC2, and MUC5AC in carcinomas of various sites: an immunohistochemical study. Am J Clin Pathol 2004;122:61-9. [Crossref] [PubMed]
- Schönleben F, Qiu W, Allendorf JD, et al. Molecular analysis of PIK3CA, BRAF, and RAS oncogenes in periampullary and ampullary adenomas and carcinomas. J Gastrointest Surg 2009;13:1510-6. [Crossref] [PubMed]
- Ahn DH, Bekaii-Saab T. Ampullary cancer: an overview. American Society of Clinical Oncology educational book 34/ASCO. Am Soc Clin Oncol Educ Book 2014.112-5. [Crossref] [PubMed]
- Oliveira-Cunha M, Hadfield KD, Siriwardena AK, et al. EGFR and KRAS mutational analysis and their correlation to survival in pancreatic and periampullary cancer. Pancreas 2012;41:428-34. [Crossref] [PubMed]


