
Up-regulation of PUM1 by miR-218-5p promotes colorectal tumor-initiating cell properties and tumorigenesis by regulating the PI3K/AKT axis
Highlight box
Key findings
• PUM1 was upregulated in T-ICs and played an essential role in the self-renewal of colorectal cells.
What is known and what is new?
• PUM1 is overexpressed in colon cancer cells with acquired resistance to cetuximab.
• Through miR-218-5p, PUM1 promotes colorectal T-IC properties and tumorigenesis by regulating the PI3K/AKT axis.
What are the implications, and what should change now?
• PUM1 is a novel biomarker of liver T-ICs and a potential target for CRC therapy.
Introduction
The prevalence of colorectal cancer (CRC) has increased globally (1). Advanced stage CRC, during the recent past, had a dismal prognosis and only a few available treatments. Patients with CRC who are ineligible for surgery are currently treated first with targeted medications (2-4). A minor percentage of CRC patients react to targeted treatments because of the genetic heterogeneity of the disease, whereas most of the patients experience no therapeutic outcomes, rather they experience highly dangerous side effects. Understanding the underlying causes of treatment resistance is therefore critical to understand. Similarly, the identification of reliable biomarkers capable of forecasting a patient’s response to medication for CRC is of key importance.
First discovered in hematological malignancies, cancer stem cells (CSCs) or tumor-initiating cells (T-ICs) have now been found in solid tumors, for instance, rectal, breast, and brain cancers (5,6). The multidirectional differentiation, self-renewal, and unlimited proliferation abilities of T-ICs, which make up a small part of the tumor tissue cells, are crucial for the development of tumors as well as for metastasis, recurrence, and chemo-resistance (7,8). T-ICs have been developed, which not only offer new theories for the clinical diagnosis and management of malignancies but also fresh perspectives on the pathophysiology of tumor growth and recurrence. Patients with CRC may experience a poor clinical outcome if their tumors contain a large T-IC population (9). Hence, thorough research on T-ICs’ regulatory mechanism and the identification of suitable intervention targets are anticipated to yield novel CRC therapeutic approaches.
PUM1, a sequence-specific RNA binding protein, participates in quite a few physiological events, for instance, the cell cycle, cell renewal, and DNA repair (10-16). Non-small-cell lung carcinoma (NSCLC), lymphocyte leukemia, ovarian cancer, and other malignancies have all been shown to involve PUM1 as an oncogene (17-21). In our earlier research, we also discovered that colon cancer cells with acquired resistance to cetuximab overexpress PUM1 (21). The findings from this work show that PUM1 exists as a novel biomarker for liver T-ICs and is thus a possible target for CRC therapy. In T-ICs, PUM1 is elevated and has a crucial role in colorectal cells’ capacity for tumorogenicity, malignant proliferation, self-renewal, and chemoresistance. We present the following article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-6/rc).
Methods
Patients and analysis
The specimens utilized in the current work were acquired from the Shanghai Fourth People’s Hospital (Shanghai, China). The Ethical Committee of the Shanghai Fourth People’s Hospital gave its approval to each of the aforementioned studies (No. 2022018-001), and they were all performed in conformity with the Declaration of Helsinki (as revised in 2013), and informed consent was obtained from all participants.
Cell lines and lentivirus
Chinese Academy of Sciences Cell Bank of Type Culture Collection (Shanghai Institute of Cell Biology of the Chinese Academy of Sciences) provided the Normal and CRC cell lines. The CRC cells were allowed to incubate at 37 ℃ with 5% CO2 and subsequently cultured separately in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% or 15% fetal bovine serum (FBS), 25 g/mL of gentamicin, 2 mM L-glutamine. GenePharma (Shanghai, China) was used to purchase all lentiviruses.
Mice and CRC induction
Shanghai Nanfang Model Biotechnology Co., Ltd. produced PUM1 TG mice (Shanghai, China). All mice were kept in ambient temperatures of 25 ℃ in a 12/12 h light-dark cycle and unrestrained supply of water and food. A single dose of diethylnitrosamine (DEN) as an intraperitoneal injection was administered to the male mice at the age of 14 days (n=6 per group, randomly assigned) in order to induce CRC. Mice were administered intraperitoneal injections of CCl4 (0.5 mL/kg in olive oil, Shanghai Macklin Biochemical Co., Ltd., Shanghai, China) at 4 weeks old. They continued this treatment for up to 12 weeks before being euthanized at 5 months. For upcoming investigations, mice livers and serum were obtained. A protocol was prepared before the study without registration.
In vivo xenograft and patient-derived xenograft (PDX) model
Male BALB/c nude mice (male, 4 weeks old) were taken from Shanghai Nanfang Model Biotechnology Co., Ltd. (Shanghai, China). As previously stated, an in vivo tumor growth experiment was carried out (22). Animal experiments were performed under a project license (No. TJBH0122101) granted by the Ethics Committee of Shanghai Tongji university, in compliance with Shanghai Fourth People’s Hospital guidelines for the care and use of animals.
Tissue dissociation and organoid culture
An organoid culture was performed as previously described (23). Different concentrations of cetuximab were added to the organs when they grew to a certain amount and size.
Spheroids assay
Three hundred single cells were seeded into 96-well ultra-low attachment microplates (Corning, USA) in serum-free DMEM/F12 (Invitrogen, USA), supplemented with B27 (1:50, Invitrogen), 20 ng/mL epidermal growth factor (EGF; Peprotech, CA, USA), 10 ng/mL basic fibroblast growth factor (bFGF; Invitrogen), and 4 mg/mL insulin (Sigma, CA, USA). The spheres were photographed and counted 7 days after seeding (primary spheres).
In vitro limiting dilution assay
Seeding of the indicated cells at various cell densities was made into 96-well ultra-low attachment culture plates and incubated for a period of 7 days. Using the LCalc version 1.1 software (Stem Cell Technologies, Inc., Vancouver, Canada) and Poisson distribution statistics, T-ICs were quantified on the basis of the frequency of wells with developing spheres
Flow cytometric analysis
To analyze the proportion of epithelial cell adhesion molecule (EpCAM) or CD24-positive liver CSCs, the colorectal cells were harvested and resuspended in the staining buffer, and then incubated with allophycocyanin (APC)-conjugated EpCAM antibody (BioLegend, Shanghai, China) or CD24 antibody (BioLegend) for 30 minutes at 4 ℃ in the dark. The cells were further washed with cold staining buffer twice and resuspended in the staining buffer containing 1 µg/mL propidium iodide (PI; BioLegend) followed by flow cytometry analysis using a Moflo XDP flow cytometer from Beckman Coulter, CA, USA. For CD24+ or EPCAM+ cell sorting, the colorectal cells (5×107) were harvested and resuspended in cold staining buffer, then incubated with antibodies against human CD24 (BioLegend) or EpCAM (BioLegend), respectively. Positively and negatively stained cells were then sorted using the Moflo XDP flow cytometer. The sorted cells from three independent experiments were subjected to a real-time polymerase chain reaction (PCR) assay.
RNA sequencing (RNA-seq)
Total RNA (PUM1 and control spheroids) was extracted using the mirVana microRNA (miRNA) Isolation Kit (Ambion, CA, USA) according to the manufacturer’s protocol. Gene ontology analysis was respectively performed using R based on the hypergeometric distribution.
Real-time PCR analysis
Total RNA was extracted from tissues or cells using TRIzol (Invitrogen) and was reverse transcribed using a Reverse Transcription System (Promega) to synthesize complementary DNA (cDNA). In the Roche Light Cycler 96 System (Roche, USA), the cDNA was mixed with SYBR Green PCR Kit (Roche) and specific primers to actuate the real-time PCR. The PCR conditions were as follows: 1 cycle at 95 ℃ for 5 minutes, followed by up to 40 cycles at 95 ℃ for 15 seconds (denaturation), 60 ℃ for 30 seconds (annealing), and 72 ℃ for 30 seconds (extension). Each experiment was repeated at least three times and the representative results were shown.
Phosphatidylinositol-3-kinase (PI3K) activity assay
PI3K activity in the immunoprecipitates was analyzed with a PI3K enzyme-linked immunosorbent assay (ELISA) kit (Echelon Biosciences, Salt Lake City, UT, USA) according to the manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed using SPSS V.18.0 (IBM, CA, USA). Data have been presented as mean ± standard deviation (SD). Comparison of three or more variables was carried out by analysis of variance (ANOVA), whereas Student’s t-test or the Mann-Whitney U-test was carried out for comparing two variables. The log-rank analysis and Kaplan-Meier method were used for the comparison of patient survival rates in various subgroups. The correlation between two variables was examined by making use of Pearson’s correlation analysis. Individual data sets underwent a different analysis. A P value <0.05 defines statistical significance.
Results
PUM1 enhances CRC oncogenesis by promoting the generation of T-ICs
PUM1 was over-expressed in a normal colorectal cell line (FHC) for elucidating the tumor driver function of PUM1 in CRC (Figure 1A). It’s interesting to note that PUM1 significantly promoted the production of T-ICs and increased the degree of expression of markers associated with T-IC in FHC cells (Figure 1A). Additionally, PUM1 caused tumor growth in a model comprising murine subcutaneous tumor and transformed colorectal cells in vitro (Figure 1B), but control cells did not (Figure 1C). PUM1 transgenic (TG) mice with specific colorectal mutations were created to further clarify the functions of PUM1 in colorectal carcinogenesis. Additionally, it was noticed that TG mice’s colorectal PUM1 expression was higher than that of wild-type (WT) mice. The control mice and PUM1-TG mice were given DEN to start the formation of tumors before receiving CCl4 twice weekly for 2 months to accelerate tumor growth. Both the control and PUM1-TG groups both produced clear colorectal tumor foci in all of the male animals. The number of tumors and maximum tumor diameters were considerably raised by PUM1’s colorectal-specific mutation (Figure 1D). Importantly TIC-associated markers, for instance, EpCAM, CD90, CD133, and CD24 had a considerably higher level of the expression in CRCs from PUM1 overexpression animals (Figure 1E), supporting the notion that PUM1 accelerated T-ICs production and facilitated liver oncogenesis.
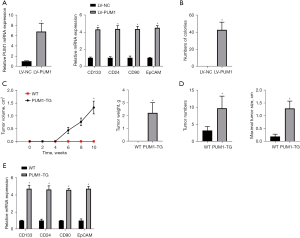
PUM1 enhances T-IC expansion in CRC
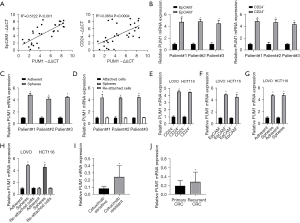
T-IC markers CD24 and EpCAM are widely used (24). PUM1 levels were favorably linked with CD24 and EpCAM expression within tumor cells derived from primary CRC tissues as demonstrated by Pearson correlation analysis (Figure 2A). We used flow cytometry sorting or sphere creation to enrich T-ICs in order to analyze PUM1 expression in T-ICs. PUM1 expression was increased in sorted EpCAM+ or CD24+ generated primary CRC cells, as seen in Figure 2B. Notably, the level of PUM1 was elevated in CRC spheres. Nevertheless, upon reattachment of the spheres, it returned to its original level (Figure 2C,2D). The outcomes from the two CRC cell lines were comparable (Figure 2E-2H). Furthermore, compared to cetuximab sensitive CRC patient tissues, the expression of PUM1 was substantially high in cetuximab resistant CRC patient tissues (Figure 2I). By chance, the expression of PUM1 was established to be considerably higher in recurrent CRC in comparison to the initial lesion (Figure 2J). PUM1 was preferentially increased in T-ICs, according to these data.
PUM1 enhances T-IC expansion by promoting PI3K/protein kinase B (AKT) activation
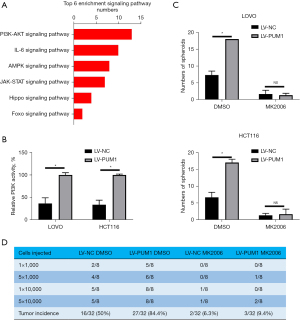
In an attempt to explain the mechanistic pathway regulating the function of PUM1 in the expansion of T-ICs, gene expression in sphere-derived PUM1 overexpression or control cells was analyzed by employing RNA-seq. Analysis of gene ontologies showed that PUM1 overexpression enriched gene sets associated with the PI3K/AKT pathway (Figure 3A). Consistently, PUM1 down expression spheroids that were generated as xenografts showed lower p-AKT levels (Figure 3B). Additionally, in colorectal T-ICs PUM1 overexpression also resulted in PI3K elevation, as demonstrated by kinase activity assays (Figure 3C). Additionally, by inhibiting AKT, PUM1 colorectal cells’ potential for self-renewal, the frequency of colorectal T-ICs, and tumorigenesis can all be improved (Figure 3D).
miR-218-5p inhibited T-ICs expansion by targeting PUM1 in CRC
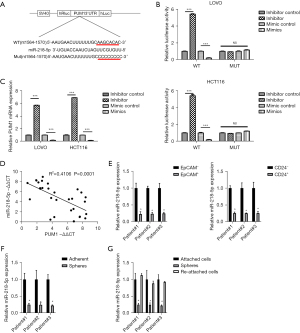
The candidate miRNA miR-218-5p that was found after searching the TargetScan database was used to further examine the upstream regulation mechanism of PUM1 in T-ICs. WT or mutant (MUT) PUM1 3'-untranslated region (3'-UTR)-coupled luciferase reporters were tested to demonstrate the binding among the PUM1 3'-UTR and miR-218-5p (Figure 4A). In CRC cell lines that had been transfected with mimics or inhibitors of this miRNA, resulting in its corresponding overexpression or downregulation, we performed functional studies of miR-218-5p (Figure S1A). Functional analysis revealed that miR-218-5p practically impedes CRC cells from proliferation, migration, as well as an invasion (Figure S1B-S1D). In CRC spheres transfected with WT PUM1 3'-UTR, miR-218-5p caused inhibition of luciferase activity, whereas a mutation in PUM1 rendered the inhibition ineffective (Figure 4B). PUM1 was further shown to be suppressed in miR-218-5p-overexpressing CRC spheres by real-time PCR (Figure 4C). In line with this, the level of expression of PUM1 and miR-218-5p in isolated primary CRC cells were inversely linked (Figure 4D). Significantly low miR-218-5p levels were observed in CRC tumor specimens in comparison to corresponding control samples (Figure S2A), and in CRC patients suffering from more advanced disease, miR-218-5p expression was notably low in comparison to the individuals with a less advanced stage of disease (Figure S2B). Furthermore, self-renewing spheroids and EpCAM+ or CD24+ primary CRC cells that were sorted showed decreased miR-218-5p expression (Figure 4E,4F). Notably, when the spheres were reattached, miR-218-5p levels returned to normal (Figure 4G). Together, the outcome demonstrates that by targeting PUM1, miR-218-5p restricted the expansion of T-ICs.
The PUM1 axis determines the cetuximab response in CRC
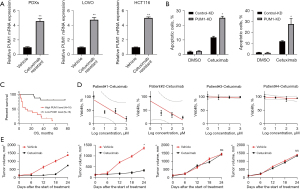
T-ICs are strongly linked to cancer’s resistance to pharmacotherapy, according to mounting data (24,25). PUM1 expression was significantly elevated in cetuximab-resistant CRC PDXs and cell lines, as illustrated in Figure 5A. PUM1 overexpression in CRC cells decreased cetuximab sensitivity in CRC cells (Figure 5B). We continue to look at the role that PUM1 plays in patient response towards cetuximab based treatment. Compared to CRC patients having a high PUM1 expression, those with low PUM1 expression had higher survival times after receiving cetuximab (Figure 5C). Next, we discovered that patient-derived organoids (PDOs) generated from tumors with high PUM1 levels were resistant to therapy with cetuximab (Figure 5D). Similar to primary CRCs, following cetuximab therapy, there was almost complete inhibition in the growth of PDXs derived from of high PUM1 carrying tumors (Figure 5E). These findings further show that PUM1 levels in tumors from patients may be a reliable indicator of cetuximab response.
Discussion
However, the incidence and death of CRC have not decreased despite improvements in research and treatment. T-ICs prevent the majority of CRC from being completely eliminated. The corresponding regulatory system for T-ICs, however, is poorly understood. In the current work, we discovered that PUM1 expression in T-ICs is downregulated due to miR-218-5p-mediated PUM1 messenger RNA (mRNA) degradation and that PUM1 inhibits T-ICs characteristics and tumorigenicity through the reduction of PI3K/AKT signaling. Our clinical studies further showed that PUM1 levels are related to patients’ benefits from cetuximab.
Despite the fact that PUM1 was increased in CRC tissues and improved CRC metastasis, its function in T-ICs has not been explored as yet. The findings of this work demonstrated that PUM1 impaired the development of T-ICs in CRC, which promoted oncogenesis. In addition, we showed that spheroid-enriched T-ICs and CD24+ or EpCAM+ cells both had higher PUM1 levels. Functional studies revealed that PUM1 improved T-IC self-renewal and growth, promoting the onset and development of CRC. PUM1 is a viable therapeutic target since, taken together, our data show that it has a significant involvement in the spread of T-ICs.
In the progression of several malignancies, including CRC, the PI3K/AKT signaling pathway is stimulated (26,27). Our RNA-seq findings demonstrated that PUM1-mediated T-ICs growth needed inhibition of the PI3K/AKT signaling pathway. According to earlier research, tissue inhibitors of metalloproteinases (TIPMs) and matrix metalloproteinases (MMPs) had a role in PUM1-mediated tumor metastasis (16). On the other hand, we suggest that PUM1 controls T-ICs expansion through the AKT signaling pathway. These findings showed the critical function of the PUM1/PI3K/AKT axis in T-ICs as well as a unique mechanism for activating hepatic T-ICs. It is anticipated that targeting the PUM1/PI3K/AKT axis may turn out to be a cutting-edge strategy for the treatment of CRC, given the significant role played by the PUM1/PI3K/AKT axis in T-ICs.
Recent evidence demonstrates that T-ICs have dysregulated miRNA expression, which aids in the growth of T-ICs (9,25). It was thus demonstrated that PUM1 downregulation in T-ICs might result from the loss or increase of miRNAs that target this gene. The luciferase reporter test identifies, MiR-218-5p as an upstream PUM1 regulator that specifically targets PUM1. A marked downregulation of miR-218-5p in T-ICs has been demonstrated in the present study and has been shown to block the PUM1/PI3K/AKT axis, which is anticipated to be the cause of both carcinogenesis and self-renewal in T-ICs. The miR-218-5p/PUM1/PI3K/AKT axis has a critical function in T-ICs, according to these findings, which also offer a new method for the activation of liver T-ICs.
There are currently few clinical options for advanced CRC patients who acquire cetuximab resistance (28-32). Therefore, it is critical to look into the causes of cetuximab resistance and to find reliable biomarkers that can forecast how well CRC patients will respond to chemotherapy. In this investigation, it was discovered that CRC cells manifested more susceptibility to cetuximab-induced growth inhibition and death due to lower PUM1 expression. Low PUM1 levels were further shown to be related to improved response and survival amongst individuals who were treated with cetuximab in the cetuximab cohort, PDO, and PDX investigations. Therefore, before choosing an appropriate course of treatment, it is recommended to assess PUM1 expression in CRC tumors for the identification of patients who can potentially benefit from cetuximab therapy. There were some limited in current version: multiple methods for cell proliferation, migration, and invasion need to be used in future.
Conclusions
Collectively, our findings highlight the crucial role of the miR-218-5p/PUM1/PI3K/AKT regulatory circuit in regulating T-IC properties, suggesting potential therapeutic targets for CRC.
Acknowledgments
Funding: This study was partially supported by the “Special Fund for Scientific Research Startup” of Shanghai Fourth People’s Hospital (No. sykyqd04401) and the Scientific Research Funding Project of Liaoning Provincial Education Department of China (No. L2019601).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-6/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-6/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-6/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-6/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present study obtained the approval of the Ethics Committee of Shanghai Fourth People’s Hospital (No. 2022018-001), and informed consent was obtained from all participants. Animal experiments were performed under a project license (No. TJBH0122101) granted by the Ethics Committee of Shanghai Tongji University, in compliance with Shanghai Fourth People’s Hospital guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rong D, Sun G, Zheng Z, et al. MGP promotes CD8(+) T cell exhaustion by activating the NF-κB pathway leading to liver metastasis of colorectal cancer. Int J Biol Sci 2022;18:2345-61. [Crossref] [PubMed]
- Li J, Huang L, Zhao H, et al. The Role of Interleukins in Colorectal Cancer. Int J Biol Sci 2020;16:2323-39. [Crossref] [PubMed]
- Fan A, Wang B, Wang X, et al. Immunotherapy in colorectal cancer: current achievements and future perspective. Int J Biol Sci 2021;17:3837-49. [Crossref] [PubMed]
- Kong L, Zhang Q, Mao J, et al. A dual-targeted molecular therapy of PP242 and cetuximab plays an anti-tumor effect through EGFR downstream signaling pathways in colorectal cancer. J Gastrointest Oncol 2021;12:1625-42. [Crossref] [PubMed]
- Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994;367:645-8. [Crossref] [PubMed]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med 2011;17:313-9. [Crossref] [PubMed]
- O'Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007;445:106-10. [Crossref] [PubMed]
- Collins AT, Berry PA, Hyde C, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005;65:10946-51. [Crossref] [PubMed]
- Li L, Tang J, Zhang B, et al. Epigenetic modification of MiR-429 promotes liver tumour-initiating cell properties by targeting Rb binding protein 4. Gut 2015;64:156-67. [Crossref] [PubMed]
- Spassov DS, Jurecic R. Cloning and comparative sequence analysis of PUM1 and PUM2 genes, human members of the Pumilio family of RNA-binding proteins. Gene 2002;299:195-204. [Crossref] [PubMed]
- Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life 2003;55:359-66. [Crossref] [PubMed]
- Gennarino VA, Singh RK, White JJ, et al. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell 2015;160:1087-98. [Crossref] [PubMed]
- Kedde M, van Kouwenhove M, Zwart W, et al. A Pumilio-induced RNA structure switch in p27-3' UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 2010;12:1014-20. [Crossref] [PubMed]
- Guan X, Chen S, Liu Y, et al. PUM1 promotes ovarian cancer proliferation, migration and invasion. Biochem Biophys Res Commun 2018;497:313-8. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Yang Y, Su X, Shen K, et al. PUM1 is upregulated by DNA methylation to suppress antitumor immunity and results in poor prognosis in pancreatic cancer. Transl Cancer Res 2021;10:2153-68. [Crossref] [PubMed]
- Şenbabaoğlu Y, Gejman RS, Winer AG, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 2016;17:231. [Crossref] [PubMed]
- Winslow S, Lindquist KE, Edsjö A, et al. The expression pattern of matrix-producing tumor stroma is of prognostic importance in breast cancer. BMC Cancer 2016;16:841. [Crossref] [PubMed]
- Fan JQ, Wang MF, Chen HL, et al. Current advances and outlooks in immunotherapy for pancreatic ductal adenocarcinoma. Mol Cancer 2020;19:32. [Crossref] [PubMed]
- Galon J, Pagès F, Marincola FM, et al. The immune score as a new possible approach for the classification of cancer. J Transl Med 2012;10:1. [Crossref] [PubMed]
- Liu Q, Xin C, Chen Y, et al. PUM1 Is Overexpressed in Colon Cancer Cells With Acquired Resistance to Cetuximab. Front Cell Dev Biol 2021;9:696558. [Crossref] [PubMed]
- Xiang DM, Sun W, Zhou T, et al. Oncofetal HLF transactivates c-Jun to promote hepatocellular carcinoma development and sorafenib resistance. Gut 2019;68:1858-71. [Crossref] [PubMed]
- Yang Y, Fan X, Ren Y, et al. SOX2-Upregulated microRNA-30e Promotes the Progression of Esophageal Cancer via Regulation of the USP4/SMAD4/CK2 Axis. Mol Ther Nucleic Acids 2021;23:200-14. [Crossref] [PubMed]
- Han T, Zhang Y, Yang X, et al. miR-552 Regulates Liver Tumor-Initiating Cell Expansion and Sorafenib Resistance. Mol Ther Nucleic Acids 2020;19:1073-85. [Crossref] [PubMed]
- Chen J, Ge X, Zhang W, et al. PI3K/AKT inhibition reverses R-CHOP resistance by destabilizing SOX2 in diffuse large B cell lymphoma. Theranostics 2020;10:3151-63. [Crossref] [PubMed]
- Zhong M, Li N, Qiu X, et al. TIPE regulates VEGFR2 expression and promotes angiogenesis in colorectal cancer. Int J Biol Sci 2020;16:272-83. [Crossref] [PubMed]
- Wang C, Chen J, Kuang Y, et al. A novel methylated cation channel TRPM4 inhibited colorectal cancer metastasis through Ca(2+)/Calpain-mediated proteolysis of FAK and suppression of PI3K/Akt/mTOR signaling pathway. Int J Biol Sci 2022;18:5575-90. [Crossref] [PubMed]
- Schaefer T, Lengerke C. SOX2 protein biochemistry in stemness, reprogramming, and cancer: the PI3K/AKT/SOX2 axis and beyond. Oncogene 2020;39:278-92. [Crossref] [PubMed]
- Zhang Y, Xia M, Jin K, et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer 2018;17:45. [Crossref] [PubMed]
- Papa E, Weller M, Weiss T, et al. Negative control of the HGF/c-MET pathway by TGF-β: a new look at the regulation of stemness in glioblastoma. Cell Death Dis 2017;8:3210. [Crossref] [PubMed]
- El Bezawy R, De Cesare M, Pennati M, et al. Antitumor activity of miR-34a in peritoneal mesothelioma relies on c-MET and AXL inhibition: persistent activation of ERK and AKT signaling as a possible cytoprotective mechanism. J Hematol Oncol 2017;10:19. [Crossref] [PubMed]
- Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018;391:1163-73. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


