
Comprehensive analysis of the correlation between GSTM1 and tumor immunity in colon cancer
Highlight box
Key findings
• GSTM1 plays an important role in the tumor immunity of COAD;
• Risk model of GSTM1-associated immunomodulators has a certain guiding value for the prognosis of COAD patients.
What is known and what is new?
• GSTM1 plays an important role in tumorigenesis and development as glutamate conjunction enzymes;
• GSTM1 is closely related to tumor immune related cells in COAD. The prognostic risk model of GSTM1-related immunomodulators was considered to be an independent prognostic factor for patients with COAD, and had good predictive accuracy for long-term survival probability.
What is the implication, and what should change now?
• GSTM1 may play a protective role in COAD by affecting tumor immunity. Further in vitro and in vivo studies were needed to verify the relevant mechanisms.
Introduction
Colon cancer is a common form of cancer with a multifactorial etiology affected by both genetic and environmental factors (1). While comprehensive surgical treatment is emphasized for colon cancer, treatment options for patients with advanced stages are very limited (2) and outcomes remain poor (3). The prognosis of colon cancer is poor, with low postoperative survival and high recurrence rates (4). New treatments to improve the prognosis of colon cancer have focused on immunotherapy strategies (5,6). Recent discoveries regarding the tumor microenvironment and the evasion of immune destruction (7) suggest immunotherapy as a precision treatment model, which may provide effective and alternative treatment approaches (8).
The immune checkpoint is a molecular hallmark to protect our immune system, which normally occurs through inhibition of T cell differentiation and proliferation to maintain the immune balance. Overexpression of the immune checkpoint molecules in tumor tissue inhibits the activation and proliferation of T cells and induces apoptosis of T cells, leading to the formation of an immunosuppressive tumor microenvironment and allowing tumor cells to escape immune monitoring (9). Immune checkpoint therapy has been shown to block inhibitory checkpoints and restore effective T cell function (10); Colon cancer was the first human tumor to be found to be immune-monitored by an adaptive immune response, and the immune system plays a complex role in colon cancer tumor immune evasion and tumor progression (11). Immune cell infiltration has been shown to have better prognostic value than classic tumor invasion criteria. While targeting the immune system with immune checkpoint inhibitors has surprising effects on some cancers, it has limited efficacy for colon cancer treatment due to the strong resistance of tumors against immune infiltration (12-14). A comprehensive understanding of colon cancer immunology and its molecular regulatory mechanism is necessary to ensure the success of immunotherapy.
Glutathione S-transferases (GSTs) are proteins that protect against oxidative stress caused by substances such as reactive oxygen species (ROS) (15). While one feature that distinguishes cancer cells from normal cells is that they can produce more ROS (16). ROS play a key role in cell signaling, cell damage, immune responses in promoting the occurrence and development of tumors (16-19). The glutathione S-transferase M1 (GSTM1) gene has been studied extensively in cancer due to its polymorphisms that are associated with tumor prognosis (20). GSTM1 can inhibit the activity of apoptosis-regulatory kinase 1 (ASK1) (21), an MAP kinase that induces the death of cytotoxic tumor cells by activating the JNK and p38 pathways (22-24). Multiple studies have indicated that a lack of GSTM1 may contribute to the occurrence of malignant tumors, including colon cancer (25,26), liver cancer (27), and breast cancer (28). Reported experimental results have suggested GSTM1 could be a potential target for colon cancer treatment; however, direct evidence is needed to reveal the role of GSMT1 in colon cancer tumor immunity to support this notion.
We investigated the potential manipulatory role of GSTM1 in colon adenocarcinoma (COAD) to propose potential GSTM1-based immunotherapy. Herein, we evaluated the expression of GSTM1 and its relationship with immune cell infiltration in 521 cases of COAD and further screened GSTM1-related immunomodulators. We then established prognostic risk signatures based on identified immunomodulators and calculated the relevant risk scores. Lastly, we constructed a nomogram by integrating the immune signature and other clinical features as prognostic biomarkers to predict the probability of long-term survival of COAD patients. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1060/rc).
Methods
Case resources and expression analysis
Transcriptional expression RNA-sequencing (RNA-seq) profile data and clinically related data were downloaded from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/). Excluding samples with incomplete data, we enrolled 14 cancer types, including at least 10 samples in the normal group. A total of 521 cases from TCGA-COAD were included, which contained 480 cancer tissues and 41 normal tissues. Furthermore, a log2 transformation was performed on the RNA-seq data in the Fragments Per Kilobase per Million (FPKM) format. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
UALCAN (http://ualcan.path.uab.edu/) is an effective online analysis and mining website for cancer data, which can be used to perform biomarker identification, expression profile analysis, and survival analysis of human genes. In this study, we conducted a comprehensive analysis of the GSTM1 protein expression data (29,30).
Correlation between GSTM1 and immune cell infiltration
ROC plotter is an online tools that can link transcriptome level data of multiple tumors with gene expression and therapeutic response (https://www.rocplot.org/) (31). The “immunotherapy” module was used to investigate the correlation between the response of 1,434 patients receiving any form of immunotherapy and the expression of GSTM1. Then we investigated the correlation between the response of patients receiving treatment and the expression of GSMT1 in various immunotherapy subgroups, including anti-programmed death 1 (PD-1) therapy, anti-programmed cell death-ligand 1 (PD-L1) therapy, and anti-cytotoxic T lymphocyte-associated antigen-4 (CTLA4) therapy. The Tumor Immune Estimation Resource (TIMER) is an online database for systematically analyzing immune cell infiltration of different cancer types (cistrome.dfci.harvard.edu/TIMER/). It contains genes from cancer histograms (TCGA) of 32 cancers focused on the correlation and survival analysis between gene expression, mutant genes, and immune infiltration abundance (32). We used the “GENE” module of TIMER to evaluate GSTM1 expression and infiltration of the six types of immune cells in COAD. Then, the “SCNA” module of TIMER was conducted to explore the correlation between somatic copy number changes and the abundance of immune infiltration in COAD.
Screening and comprehensive analysis of GSTM1-related immunomodulators
We extracted immunomodulators that were significantly correlated with GSTM1 through TISIDB (http://cis.hku.hk/TISIDB/) (33). Furthermore, in this study, we used the STRING database (http://string-db.org/) to construct a protein-protein interaction (PPI) network by GSTM1-related immunomodulators (high confidence, 0.700). We aimed to understand the working principle of each protein, the reaction mechanism of biological signals and energy substance metabolism under pathological conditions, and the functional relationship between proteins (34). Additionally, functional enrichment analysis of the co-expression gene set was performed with the clusterProfiler package (V 3.14.3) and further visualized with the ggplot2 package (V3.3.3) (35).
Establishment and evaluation of a prognostic risk model
We further determined a prognostic multiple immune gene signature out of the GSTM1-associated immunomodulators. Single-factor Cox analysis was used to identify immune prognostic-related genes (P<0.05), and multivariate Cox regression analysis was used to finally develop an immune-related prognostic risk model (36). Patients were divided into high- and low-risk groups using the median risk score as the cutoff value. The Kaplan-Meier survival curve was performed for the risk model prognosis analysis. The prognostic accuracy of the risk score was determined by the time-dependent receiver operating characteristic (ROC) curve using “time ROC” packages. The risk curve of the model was used to assess the significance of the difference in survival between the high- and low-risk groups (37). Additionally, multivariate Cox analysis was performed after adjusting for age, sex, and stage to verify the independent prognostic implications of the risk score.
We assessed the individual prognosis of TCGA-COAD patients using nomogram, which is based on multivariate regression analysis and integrates multiple predictors to express the relationship between variables in the prediction model (38). In this study, based on the result of multifactor regression analysis, we used the “RMS” R package [6.2-0 version] to construct a nomogram to predict the possible overall survival (OS) of the individual patients at one year, three years, and five years, including multiple predictors such as gender, age, Tumor-Node-Metastasis (TNM) stage, and risk score. and the relationships between these variables were transformed into a visual graph to make the results of the predictive model more readable.
Statistical analysis
Statistical analyses were conducted by R V 4.0.5 and V 3.6.3. Visualization was performed using ggplot2 (V 3.3.3). The Mann-Whitney U and Z tests were performed to identify the GSTM1 expression level. Survival curves were generated using the Kaplan-Meier method. The correlational analysis of gene expression was performed by the Spearman method. Univariate and multivariate analyses were conducted using Cox regression models to determine independent prognostic factors. A P value <0.05 was considered statistically significant.
Results
Downregulation of GSTM1 mRNA and protein expression in COAD
We searched and analyzed a dataset of 14 cancers in the TCGA database with the filter condition that the normal tissues of each cancer contained at least 10 samples, and the mRNA expression level of GSTM1 in different types of cancer was estimated. As shown in Figure 1A, compared with normal tissues, the expression of GSTM1 was significantly downregulated in COAD, gastric adenocarcinoma, prostate cancer, thyroid cancer, and endometrial cancer. These results are consistent with previous meta-analysis results (25,27,39,40).
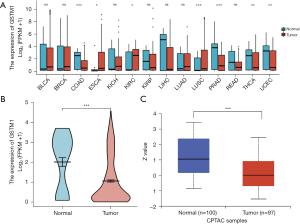
We further analyzed the mRNA and protein expression of GSTM1 in COAD using the TCGA and HPA databases. As shown in Figure 1B, mRNA expression levels of GSTM1 in COAD tissues (n=480) were remarkably lower than those in adjacent tissues (n=41). To carry out a comprehensive analysis of GSTM1 protein expression, we used the UALCAN online tool. The results showed that the protein expression Z-value of GSTM1 was decreased in COAD tissues (Figure 1C). In summary, these results provide evidence for the downregulation of GSTM1 in COAD.
GSTM1 is correlated to the infiltration of immune cells
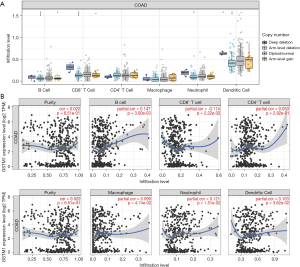
Further, we tried to determine whether GSTM1 expression is related to immune cell infiltration in COAD. For this purpose, we conducted a correlation analysis through the TIMER “GENE” and “SCNA” modules. As shown in Figure 2A, the infiltration levels of B cells, CD8+ T cells, neutrophils, and dendritic cells were decreased with the chromosome arm-level deletion. In addition, GSTM1 expression positively correlated with B cells (r=0.147, P=0.003), macrophages (r=0.099, P=0.047), neutrophils (r=0.121, P=0.015), and dendritic cells (r=0.103, P=0.038) (Figure 2B).
GSTM1 expression is related to immunomodulators
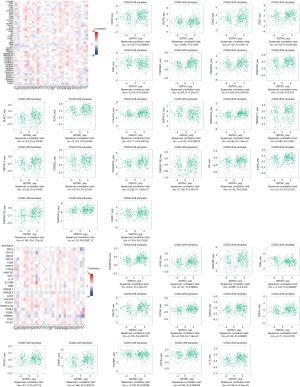
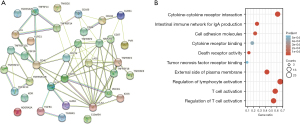
We assessed the relationship between GSTM1 expression and immunomodulators based on the TISIDB baseline. Our results identified 23 immunostimulators and 13 immunoinhibitors that were positively or negatively associated with GSTM1 expression in COAD (Figure 3). Based on these 36 GSTM1-associated immunomodulators, we constructed a PPI network using the STRING database. A strong correlation (r=0.7) of the 36 GSTM1-associated immunomodulators is shown in Figure 4A. At the same time, a pathway enrichment analysis for the 36 GSTM1-associated immunomodulators was performed, and our results demonstrated that some crucial related pathways, including T cell activation, regulation of T cell activation, and regulation of lymphocyte activation, were related to GSTM1-mediated immune events (Figure 4B).
Establishment of a GSTM1-related immune prognostic risk model
To study the prognostic value of GSTM1-associated immunomodulators in COAD, we included the 36 GSTM1-associated immunomodulators in a stepwise multivariate Cox regression analysis and obtained an optimal 2-gene immune-related prognostic signature (TNFRSF13C and TNFRSF25) in TCGA-COAD patients (Figure 5A). In this prognostic risk model, the risk score for each patient can be calculated following the proposed formula: TNFRSF13C expression level × its coefficient (0.466) + TNFRSF25 expression level × its coefficient (0.253). The TCGA-COAD patients were assigned into high- or low-risk groups based on the median value of the risk score. Afterward, we performed a Kaplan-Meier survival analysis based on the log-rank test; the results showed that the OS of high-risk patients was worse than that of low-risk patients (P=0.023) (Figure 5B). Next, we used the time-dependent ROC curve to assess the prediction accuracy of the risk model, and the area under the curve (AUC) values of the risk score and stage were 0.496 and 0.675, respectively. The AUC value was 0.747 when the risk score was combined with stage, suggesting a better prediction performance (Figure 5C). Then, we ranked each patient’s risk score in ascending order, and the risk distribution map was plotted. As shown in Figure 5D, the expression of the high-risk genes TNFRSF13C and TNFRSF25 were upregulated with the increasing risk score, and fewer deaths in the low-risk group were observed. In addition, using univariate and multivariate Cox regression, the risk score was identified as an independent prognostic factor for TCGA-COAD patients when adjusted for age and stage [hazard ratio (HR) =2.068, 95% confidence interval (CI): 1.628–2.626, P<0.001] (Figure 5E).
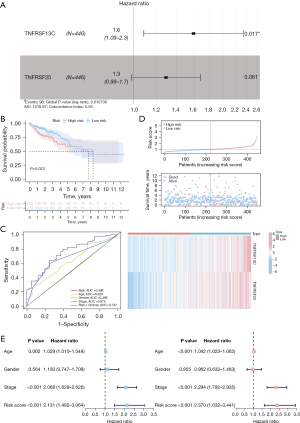
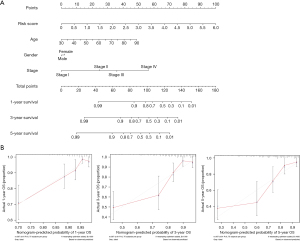
Finally, we constructed a prognostic nomogram by integrating the risk scores with other key clinical features, such as age and grade. Our constructed nomogram is a good tool for an accurate clinical prognostic assessment with a concordance index (C-index) value of 0.583 (Figure 6A). Moreover, the calibration curve showed that the probability predicted by the nomogram (gray line) had a good overlap with the ideal reference line (red line) for 3- and 5-year survival rates and especially for the 1-year survival rate (Figure 6B).
Discussion
In this study, we used different databases to study the expression differences of the GSTM1 gene in cancer and normal tissues. We showed that expression of the GSTM1 gene in gastrointestinal tumors such as colon cancer, gastric adenocarcinoma, prostate cancer, thyroid cancer, and endometrial cancer was low compared to that in normal tissues. In addition, by further studying the mRNA dataset and protein expression of colon cancer, we found that the expression of the GSTM1 gene was significantly downregulated in colon cancer. Another major finding of this study is that GSTM1 expression in COAD was related to the infiltration of a variety of immune cells. The TIMER analysis showed that the decreased copy number of GSTM1 may lead to a decrease in B cells and that GSTM1 expression was positively correlated with B cell infiltration. In addition, we constructed two GSTM1-related immunomodulator risk prognostic models (TNFRSF13C and TNFRSF25), in which TNFRSF13C is a member of the tumor necrosis factor (TNF) receptor superfamily. It has been reported that TNFRSF13C is also involved in the development of B lymphocytes and the survival of mature B cells (41-43). Moreover, TNFRSF13C is an attractive target for B-cell lymphoma (44). The evidence further emphasizes the importance of GSTM1-associated B cell-mediated immunity in COAD. Additionally, our findings displayed a positive correlation between GSTM1 expression and macrophage infiltration. It has been well established that macrophages are the key components involved in inflammatory and tumor immune responses (45-47). Furthermore, research on therapy for tumor-associated macrophages suggests that various components of macrophages may become targets for new tumor treatments (48).
In addition, our study demonstrated that GSTM1-associated immunomodulators are involved in several crucial immune processes, including T cell activation, regulation of T cells, and lymphocyte activation. Lymphocytes are important immunocompetent cells in the immune system (49). The signal transduction and molecular basis of their activation process are extremely complex, and these immune processes have been considered important hallmarks for the prevention and control of cancer (50). Especially, T cell activation is a key and indispensable step for the anti-tumor immune response, and the effect mechanism of PD-L1 is also based on T cell activation (40). Moreover, PD-L1 can bind to the programmed cell death protein 1 of T cells, resulting in the inhibition of T-cell activity and impairment of anticancer T-cell immunity (51). Indeed, we also observed that the expression level of GSTM1 in patients receiving anti PD-L1 treatment and with response were higher than that those without response when investigating the response of GSTM1 and immunotherapy (Figure S1). Based on the effect of GSTM1 on immune cells and immune-related processes, we conclude that GSTM1 might be a therapeutic target in COAD.
In recent years, several immune-related gene signatures have been identified and used for the prognosis prediction of colon cancer (52). For instance, Li et al. constructed a ten-gene immune-related signature that reflected the immune microenvironment and survival rates in colon cancer (53). Zhang et al. reported an immune-paired gene signature based on several public databases, which showed an independent prognostic role in colon cancer (54). We identified a two-gene immune-related signature (TNFRSF13C and TNFRSF25) and established an immune-related prognostic risk model based on GSTM1-associated immunomodulators following a stepwise multivariate Cox regression analysis. To further study the clinical value of the immune-related prognostic risk model, we evaluated the relationship between the model and the OS of patients. We demonstrated that patients with high-risk scores had a worse prognosis. Multivariate Cox analysis further confirmed that the risk score of the model was an independent predictor for COAD patients. In addition, we combined the risk score with other key clinical features to construct a prognostic nomogram, which quantitatively predicts the individual risk of COAD. Moreover, the 1-year survival probability calibration curve demonstrated a good agreement between the survival probability predicted by the nomogram and the actually observed survival probability. These findings indicate that GSTM1-related 2 immunomodulators may be a good prognostic hallmark for COAD patients.
Conclusions
The combination of GSTM1 and immune cell infiltration assessments is expected to allow for a more accurate evaluation of prognosis. However, the above results are bioinformatics analyses based on public data, which is a limitation of this study. It is necessary to carry out further experiments for verification. Our research provides evidence that GSTM1 is closely correlated to tumor immunity. Furthermore, our identified GSTM1-related risk signature is a potential novel prognostic biomarker for prognosis in colon cancer.
Acknowledgments
Funding: This work was supported by the General Project of Chongqing Natural Science Foundation (No. cstc2021jcyj-msxmX0153 to LLP; No. cstc2021jcyj-msxmX0112 to YXW; No. cstb2022nscq-msx1005 to HTG), the Science and Technology Research Program of Chongqing Municipal Education Commission (No. KJQN201900401 to YHT), and the Kuanren Talents Program of The Second Affiliated Hospital of Chongqing Medical University (No. kryc-yq-2110 to HTG).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1060/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1060/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019;16:713-32. [Crossref] [PubMed]
- Crutcher MM, Baybutt TR, Kopenhaver JS, et al. Emerging drug targets for colon cancer: A preclinical assessment. Expert Opin Ther Targets 2022;26:207-16. [Crossref] [PubMed]
- Ferrer-Mayorga G, Gómez-López G, Barbáchano A, et al. Vitamin D receptor expression and associated gene signature in tumour stromal fibroblasts predict clinical outcome in colorectal cancer. Gut 2017;66:1449-62. [Crossref] [PubMed]
- Kumar S, Burney IA, Zahid KF, et al. Colorectal Cancer Patient Characteristics, Treatment and Survival in Oman--a Single Center Study. Asian Pac J Cancer Prev 2015;16:4853-8. [Crossref] [PubMed]
- Fan A, Wang B, Wang X, et al. Immunotherapy in colorectal cancer: current achievements and future perspective. Int J Biol Sci 2021;17:3837-49. [Crossref] [PubMed]
- Kishore C, Bhadra P. Current advancements and future perspectives of immunotherapy in colorectal cancer research. Eur J Pharmacol 2021;893:173819. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest 2015;125:3335-7. [Crossref] [PubMed]
- Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350-5. [Crossref] [PubMed]
- Alsaab HO, Sau S, Alzhrani R, et al. PD-1 and PD-L1 Checkpoint Signaling Inhibition for Cancer Immunotherapy: Mechanism, Combinations, and Clinical Outcome. Front Pharmacol 2017;8:561. [Crossref] [PubMed]
- Wang K, Karin M. Tumor-Elicited Inflammation and Colorectal Cancer. Adv Cancer Res 2015;128:173-96. [Crossref] [PubMed]
- Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960-4. [Crossref] [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- Foroutan M, Molania R, Pfefferle A, et al. The Ratio of Exhausted to Resident Infiltrating Lymphocytes Is Prognostic for Colorectal Cancer Patient Outcome. Cancer Immunol Res 2021;9:1125-40. [Crossref] [PubMed]
- Strange RC, Spiteri MA, Ramachandran S, et al. Glutathione-S-transferase family of enzymes. Mutat Res 2001;482:21-6. [Crossref] [PubMed]
- Prasad S, Gupta SC, Tyagi AK. Reactive oxygen species (ROS) and cancer: Role of antioxidative nutraceuticals. Cancer Lett 2017;387:95-105. [Crossref] [PubMed]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 2004;55:373-99. [Crossref] [PubMed]
- Bekeschus S, Emmert S, Clemen R, et al. Therapeutic ROS and Immunity in Cancer-The TRIC-21 Meeting. Cancers (Basel) 2021.
- Sun L, Wang X, Saredy J, et al. Innate-adaptive immunity interplay and redox regulation in immune response. Redox Biol 2020;37:101759. [Crossref] [PubMed]
- McIlwain CC, Townsend DM, Tew KD. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene 2006;25:1639-48. [Crossref] [PubMed]
- Cho SG, Lee YH, Park HS, et al. Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J Biol Chem 2001;276:12749-55. [Crossref] [PubMed]
- Ichijo H, Nishida E, Irie K, et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science 1997;275:90-4. [Crossref] [PubMed]
- Saitoh M, Nishitoh H, Fujii M, et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J 1998;17:2596-606. [Crossref] [PubMed]
- Dorion S, Lambert H, Landry J. Activation of the p38 signaling pathway by heat shock involves the dissociation of glutathione S-transferase Mu from Ask1. J Biol Chem 2002;277:30792-7. [Crossref] [PubMed]
- Economopoulos KP, Sergentanis TN. GSTM1, GSTT1, GSTP1, GSTA1 and colorectal cancer risk: a comprehensive meta-analysis. Eur J Cancer 2010;46:1617-31. [Crossref] [PubMed]
- Ay A, Gulyasar T, Alkanli N, et al. Investigation of the relationship between GSTM1 gene variations and serum trace elements, plasma malondialdehyde levels in patients with colorectal cancer. Mol Biol Rep 2021;48:6911-21. [Crossref] [PubMed]
- Wang B, Huang G, Wang D, et al. Null genotypes of GSTM1 and GSTT1 contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. J Hepatol 2010;53:508-18. [Crossref] [PubMed]
- Dos Santos SP, Morissugui SS, Gimenez Martins APD, et al. Evaluation of molecular markers GSTM1 and GSTT1 and clinical factors in breast cancer: case-control study and literature review. Xenobiotica 2021;51:1326-34. [Crossref] [PubMed]
- Edwards NJ, Oberti M, Thangudu RR, et al. The CPTAC Data Portal: A Resource for Cancer Proteomics Research. J Proteome Res 2015;14:2707-13. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Fekete JT, Győrffy B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int J Cancer 2019;145:3140-51. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39:D561-8. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One 2013;8:e74250. [Crossref] [PubMed]
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000;56:337-44. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Song L, Yang C, He XF. Individual and combined effects of GSTM1 and GSTT1 polymorphisms on colorectal cancer risk: an updated meta-analysis. Biosci Rep 2020;40:BSR20201927. [Crossref] [PubMed]
- Mo Z, Gao Y, Cao Y, et al. An updating meta-analysis of the GSTM1, GSTT1, and GSTP1 polymorphisms and prostate cancer: a HuGE review. Prostate 2009;69:662-88. [Crossref] [PubMed]
- Gross JA, Dillon SR, Mudri S, et al. TACI-Ig neutralizes molecules critical for B cell development and autoimmune disease. impaired B cell maturation in mice lacking BLyS. Immunity 2001;15:289-302. [Crossref] [PubMed]
- Li YJ, Jiang WQ, Rao HL, et al. Expression of BAFF and BAFF-R in follicular lymphoma: correlation with clinicopathologic characteristics and survival outcomes. PLoS One 2012;7:e50936. [Crossref] [PubMed]
- Sevdali E, Block Saldana V, Speletas M, et al. BAFF receptor polymorphisms and deficiency in humans. Curr Opin Immunol 2021;71:103-10. [Crossref] [PubMed]
- Qin H, Wei G, Sakamaki I, et al. Novel BAFF-Receptor Antibody to Natively Folded Recombinant Protein Eliminates Drug-Resistant Human B-cell Malignancies In Vivo. Clin Cancer Res 2018;24:1114-23. [Crossref] [PubMed]
- Mantovani A, Sozzani S, Locati M, et al. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol 2002;23:549-55. [Crossref] [PubMed]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012;122:787-95. [Crossref] [PubMed]
- Chen D, Zhang X, Li Z, et al. Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages. Theranostics 2021;11:1016-30. [Crossref] [PubMed]
- Gabrilovich DI. Myeloid-Derived Suppressor Cells. Cancer Immunol Res 2017;5:3-8. [Crossref] [PubMed]
- Gautam N, Das S, Kar Mahapatra S, et al. Age associated oxidative damage in lymphocytes. Oxid Med Cell Longev 2010;3:275-82. [Crossref] [PubMed]
- Campian JL, Ye X, Gladstone DE, et al. Pre-radiation lymphocyte harvesting and post-radiation reinfusion in patients with newly diagnosed high grade gliomas. J Neurooncol 2015;124:307-16. [Crossref] [PubMed]
- Roemer MG, Advani RH, Ligon AH, et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J Clin Oncol 2016;34:2690-7. [Crossref] [PubMed]
- Lu Z, Chen J, Yan J, et al. A 13-Immune Gene Set Signature for Prediction of Colon Cancer Prognosis. Comb Chem High Throughput Screen 2021;24:1205-16. [Crossref] [PubMed]
- Li M, Wang H, Li W, et al. Identification and validation of an immune prognostic signature in colorectal cancer. Int Immunopharmacol 2020;88:106868. [Crossref] [PubMed]
- Zhang Q, Feng Z, Zhang Y, et al. Identification and Verification of a 17 Immune-Related Gene Pair Prognostic Signature for Colon Cancer. Biomed Res Int 2021;2021:6057948. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


