
Circ_0000799 promotes proliferation and invasion in colorectal cancer and epithelial-mesenchymal transition
Highlight box
Key findings
• Circ_0000799 is up-regulated in colorectal cancer (CRC) tissues and cells.
What is known and what is new?
• It has been found that non-coding RNAs play an important regulatory role in living cells and diseases. Circular RNAs (circRNAs) are newly discovered non-coding RNAs with circular structures.
• We explore the effects and mechanisms of circ_0000799 expression on CRC cells to provide experimental data and a basis for the search for diagnostic and therapeutic targets for CRC.
What is the implication, and what should change now?
• Circ_0000799 can be used as a novel biomarker for CRC diagnosis and has the potential to be a novel therapeutic target for CRC.
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide. In 2017, the number of new CRC patients was 1.8 million; it has the third highest incidence among cancers and the characteristics of high metastasis and high recurrence rate (1). Currently, CRC ranks the 4th in incidence rate and 5th in mortality of malignant tumors in China (2). The number of deaths from CRC was 890,000 in 2017, and its mortality is the second highest among cancer-related deaths. The 5-year relative survival rate of CRC patients is 65%. If CRC is diagnosed in the pre-metastatic stage, the 5-year relative survival rate of patients can be increased to 90% (3), indicating that early and accurate diagnosis is critical for the treatment of CRC. At present, colonoscopy and blood markers are clinically used to diagnose CRC (4). Blood markers include cancer antigen (CA)19-9, CA724, carcinoembryonic antigen (CEA), and so on. However, both the sensitivity and specificity of these diagnostic methods are unsatisfactory. In addition, many factors affect the occurrence and development of CRC, such as age, genetics, and environmental factors (5). CRC is mainly manifested in hematochezia caused by rectal bleeding, iron deficiency, anemia, abdominal pain, weight loss, and loss of appetite (6). Clinically, the treatment of the disease is usually based on surgery and supplemented with chemotherapy and radiotherapy. However, the prognosis of CRC patients remains unsatisfactory (7). With the in-depth study of CRC, some researchers have proposed molecular targeted therapy, such as vascular endothelial growth factor (VEGF) tyrosine kinase inhibitors and anti-epidermal growth factor receptor (EGFR) monoclonal antibodies (8). Although the disease can be improved by targeting these molecules, the effect is limited (8). Therefore, there is an urgent need to find more effective therapeutic targets to improve the diagnostic efficiency and treatment of CRC.
In recent years, with the deepening of the research on non-coding RNAs, it has been found that non-coding RNAs play an important regulatory role in living cells and diseases. Circular RNAs (circRNAs) are newly discovered non-coding RNAs with circular structures. Different from other non-coding RNAs, the circular structure contributes to the stability of circRNA, so this circRNA is not easily degraded (9). It has been confirmed that circRNAs are involved in cancer-related physiological and pathological processes, for example, cell proliferation, migration, invasion, cell cycle, and carcinogenesis (10). Zeng et al. found that a significantly up-regulated expression of circHIPK3 in CRC tissues and cell lines can be used as a marker for the prognosis of CRC patients and this circRNA can improve the proliferation and metastasis of CRC cells by down-regulating miR-7 (11). Li et al. demonstrated that circDDX17 was significantly down-regulated in CRC tissues, indicating that this circRNA may have an antitumor effect on CRC (12). The above studies suggest that circRNAs may play a crucial role in the development of CRC. However, due to the variety and complex function of circRNAs, many CRC related circRNAs are still to be discovered. Circ_0000799 (circ-BPTF) is a circRNA derived from bromodomain PHD finger transcription factor (BPTF) exons. Bi et al. revealed that the expression of circ_0000799 was increased in bladder cancer tissues and cell lines, and this circRNA could promote the progression of bladder cancer by affecting the activity of the miR-31-5p/RAB27A signaling axis (13). These results suggest that circ_0000799 may be associated with promoting the development of cancer. However, there is currently no study on the relationship between circ_0000799 and CRC occurrence and development. Thus, by transfecting CRC cells with circ_0000799, we explore the effects and mechanisms of circ_0000799 expression on CRC cells to provide experimental data and a basis for the search for diagnostic and therapeutic targets for CRC. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1176/rc).
Methods
Collection of clinical tissues
CRC tissue samples and CRC-adjacent normal tissues (Normal group) were collected from patients treated in The Fifth Affiliated Hospital of Harbin Medical University between June 2017 and December 2020. The study was conducted according to the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Ethics Committee of The Fifth Affiliated Hospital of Harbin Medical University (No. KY-2022-019), and all of the patients provided their written informed consent.
Cell culture and transfection
Human normal colon epithelial cells (FHC group) and CRC cell lines (HCT116, HT29, SW480, and SW620) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in complete Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS; Gibco, Waltham, MA, USA), 100 mg/mL penicillin, and 10 mg/mL streptomycin (Gibco) in an incubator at 37 ℃ with 5% CO2 (Thermo Fisher, Waltham, MA, USA). Circ_0000799 overexpressed plasmids and interfering fragments as well as their negative controls (NCs) were designed and synthesized by GenePharma (Shanghai, China). The sequence of small interfering (si)-circ0000799 is: AAGUAGCAGGUCAGGUGTT. According to the Lipofectamine 3000 (Thermo Fisher) transfection step, HCT116 and SW480 cells were transfected with negative plasmid (vector group), circ_0000799 overexpression plasmid (circ_0000799 group), siRNA-NC (si-NC group), and siRNA-circ_0000799 (si-circ_0000799 group), respectively.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Extraction of total RNA from cancerous tissues and cells by TRizol method, and nuclear and cytoplasmic RNA was isolated from HCT116 and SW480 cells using a nucleoplasmic isolation kit. Subsequently, use NanoDrop (Thermo Fisher) to detect the concentration and purity of RNA, and complementary DNA (cDNA; Thermo Fisher) was prepared according to random primer reverse transcription kit. The expression levels of circ_0000799, miR-647, and miR-1243 were detected according to the instructions of the SYBR GREEN kit (TaKaRa, Shiga, Japan), and RNU6B (U6) was taken as the internal reference. A total of 6 replicates were set up in the experiment. The experimental data obtained by qRT-PCR were used to calculate relative gene expression levels by the 2−ΔΔCt method. The primer sequences were as follows: circ_0000799, forward (F): 5'-UCAGCUACAAAUACAGCAGCCAC-3', reverse (R): 5'-AGUCGAUGUUUAUGUCGUCGGUG-3'; miR-647, F: 5'-CUUCCUCACUCACGUCGGUG-3', R: 5'-GAAGGAGUGAGUGCAGCCAC-3'; miR-1243, F: 5'-AACTGGATCAATTATA-3', R: 5'-GTGCAGGGTCCGAGGT-3'; U6, F: 5'-CTCGCTTCGGCAGCACATA-3', R: 5'-AACGCTTCACGAATTTGCGT-3'.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
HCT116 and SW480 cells in the logarithmic growth phase after treatment were inoculated in 96-well plates at a density of 3×103 cells/well and cultured for 24, 48, and 72 hours, respectively. Then, 20 µL of 5 mg/mL MTT solution was added to each group of cells and cultured for another 4 hours in the incubator. After the supernatant was aspirated, the samples were added with 150 µL of dimethyl sulfoxide (DMSO) and then shaken for 15 minutes. The absorption at 570 nm in each test well was measured using a microplate reader.
Colony formation assay
The treated HCT116 and SW480 cells were digested with 0.25% trypsin and resuspended in DMEM with 0.35% agarose and L-15 complete media. Then, the resuspended cells were seeded in 6-well plates containing 0.6% agarose at a density of 1×105 cells/well, and incubated in CO2 incubator at 37 ℃. After colony formation was observed, the cells were stained with 0.1% crystal violet solution. Finally, the colonies were photographed with an inverted microscope and counted.
Transwell assays
Cell migration and invasion assays were performed using transwell inserts. In the migration assay, 100 µL of transfected cells were seeded in the upper chamber of the transwell inserts, and 700 µL of medium containing 20% FBS was added to the lower chamber. The transwell insert was taken out after 24 hours of incubation and washed 3 times with phosphate-buffered saline (PBS). After the cells were fixed in 20% methanol for 30 minutes, they were stained with 0.1% crystal violet for 12 hours. The staining of the cells was observed under an inverted microscope. Invasion assays were similar to the migration assays except that Matrigel [Becton, Dickinson, and Co. (BD) Biosciences, Franklin Lakes, NJ, USA] was coated in the center of the upper chamber before cell seeding. In the migration and invasion assays, three areas were randomly selected for photographic and quantitative analysis.
Dual luciferase reporter assay
The prediction of microRNAs (miRNAs) bound to circ_0000799 was carried out by using CircInteractome (https://circinteractome.nia.nih.gov/index.html), and circBank (http://www.circbank.cn/). The wild-type (WT) and mutant (MUT) sequence of circ_0000799/miR-647 binding site was inserted into downstream of the firefly luciferase gene to construct an expression vector (Promega, Madison, WI, USA). By using Lipofectamine 2000 transfection reagent, miR-647 mimics/mimics NC was mixed with pmirGLO-circ_0000799-WT/MUT recombinant plasmid and then transfected into 293T cells. After 48 hours of transfection, luciferase activity was measured according to the instructions of the dual-luciferase reporter assay kit.
Western blot
The total protein of cells was extracted by cell lysate and the protein concentration was determined with the bicinchoninic acid (BCA) protein quantitative kit (Abcam, Cambridge, UK). Subsequently, 20 µg of protein with 2× loading buffer was boiled for denaturation. Proteins were isolated using sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF) membrane. Afterward, the membrane was blocked by 5% skim milk powder for 1 hour. The membrane was incubated with primary antibody anti-Vimentin antibody (ab92547, Abcam, USA), anti-N-cadherin antibody (ab18203, Abcam, USA), anti-E-cadherin antibody (ab40772, Abcam, USA), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (ab8245, Abcam, USA), at 4 ℃ overnight. The next day, the membrane was incubated with secondary antibodies goat anti-rabbit IgG H&L horseradish peroxidase (HRP) (ab205718, Abcam, USA), goat anti-mouse IgG H&L HRP (ab205719, Abcam, USA) for 1 hour at ambient temperature after washing the membrane 3 times. After incubation, the membrane was rinsed 3 times again. Then, enhanced chemiluminescence reagents were used to visualize the proteins. Photographs were taken using a gel imaging system. Image J analysis software (National Institutes of Health, Bethesda, MD, USA) was used to quantify the gray levels of bands in the western blot image, and GAPDH was used as an internal reference to calculate the relative protein expression.
Statistics analysis
Statistics analysis was conducted by SPSS 26.0 (IBM Corp., Armonk, NY, USA). Measurement data were expressed by mean ± standard deviation (SD). Differences between or among groups were compared using Students’ t-test or one-way analysis of variance (ANOVA). The correlation between circ_0000799 expression and miR-647 in CRC tissues was analyzed using Pearson correlation analysis. Statistical significance was defined at P<0.05.
Results
Circ_0000799 is up-regulated in CRC tissues and cells
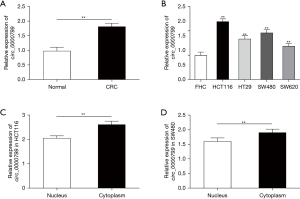
Compared with the Normal group, the expression of circ_0000799 was significantly increased in CRC tissues (Figure 1A, P<0.01). Compared with the FHC group, the expression of circ_0000799 was significantly up-regulated in the CRC cell lines (HCT116, HT29, SW480, SW620) (Figure 1B, P<0.05). This result suggested that circ_0000799 may be involved in the progression of CRC. Further experiments confirmed that circ_0000799 was mainly expressed in the cytoplasm (Figure 1C,1D, P<0.05).
Knockdown of circ_0000799 suppresses the proliferation and viability of CRC cells
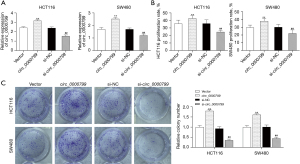
To investigate the role of circ_0000799 in CRC, we interfered with or overexpressed circ_0000799 in HCT116 and SW480 cells. As shown in Figure 2A, the expression of circ_0000799 was significantly increased in the circ_0000799 group compared with the vector group; the expression of circ_0000799 was notably reduced in the si-circ_0000799 group compared with the si-NC group (P<0.05). Collectively, the results of MTT and colony formation assay showed that the proliferation rate and cell viability of the circ_0000799 group were significantly increased compared with the vector group; the proliferation rate and cell viability of the si-circ_0000799 group were significantly decreased compared with the si-NC group (Figure 2B,2C).
Knockdown of circ_0000799 suppresses the invasion and migration of CRC cells and the epithelial-mesenchymal transition (EMT) process
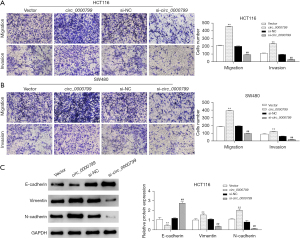
We aimed to further assess the effect of circ_0000799 on CRC cell invasion and migration. In HCT116 and SW480 cells, the migration and invasion of CRC cells were significantly promoted after overexpression of circ_0000799 while inhibited after interference with circ_0000799 expression (Figure 3A,3B, P<0.05). Furthermore, western blot results revealed that the expression of vimentin and N-cadherin were significantly increased, whereas the protein level of E-cadherin was significantly decreased in HCT116 cells in the circ_0000799 group compared with the vector group (Figure 3C). However, knockdown of si-circ_0000799 resulted in opposite changes in the expression of the above proteins.
Circ_0000799 serves as a sponge of miR-647
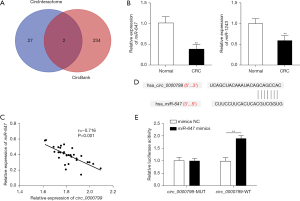
We further analyzed the potential molecular mechanism of circ_0000799 in CRC. We predicted interacting miRNAs of circ_0000799 in CircInteractome and circBank databases predicted the miRNAs which can potentially target the circ_0000799, and finally found miR-647 and miR-1243 as the potential ones based on the intersection of the two databases (Figure 4A). The qRT-PCR analysis revealed that the expression levels of miR-647 and miR-1243 were notably reduced in CRC tissues, and the former was lower (Figure 4B, P<0.05). Pearson correlation analysis showed that the expressions of circ_0000799 and miR-647 were negatively correlated in CRC (Figure 4C). The binding site of circ_0000799 to miR-647 was predicted by CircInteractome (Figure 4D). Subsequently, a dual-luciferase reporter assay showed that miR-647 mimics significantly reduced circ_0000799-WT 3'-untranslated region (3'-UTR) luciferase activity, but had no effect on the luciferase activity of circ_0000799-MUT plasmid (Figure 4E). Collectively, circ_0000799 could bind to miR-647 and negatively regulate its expression.
Discussion
With the deepening of research on circRNAs, studies have shown that circRNAs play crucial roles in cancer occurrence and development (14). At the same time, due to the special ring structure, circRNAs seem to be more suitable as biomarkers for disease. Indeed, many circRNAs have been recognized as biomarkers for cancers, including gastric cancer, liver cancer, bladder cancer, and so on (15). However, representative circRNAs acting as indicators of other diseases have not been fully mined, including CRC. In previous studies, the expression of hsa_circ_0000069 and hsa_circ_0007534 was significantly up-regulated in CRC tissues compared with adjacent non-tumor tissues (16,17), yet the expression of circ_0026344 and hsa_circ_001988 was notably reduced (18,19). A study also showed that elevated expression of circ_0000745 in CRC tissues (20). All of the above circRNAs can serve as biomarkers for CRC diagnosis. However, some studies have found that hsa_circ_0000069 is also up-regulated in cervical cancer tissues (21), and hsa_circ_0007534 is up-regulated in osteosarcoma and cervical cancer tissues (22,23). It is suggested that the change of single circRNA expression is not specific and is affected by many factors. Therefore, the diagnosis rate can be improved if we use the change of multiple circRNAs expressions as the diagnostic basis. In the results of this assay, the expression level of circ_0000799 was significantly increased in CRC tissues and cells. Bi et al. found that circ_0000799 (circ-BPTF) was also up-regulated in bladder cancer tissues (13). Collectively, it is indicated that circ_0000799 is closely related to the development of cancer, and can be used as a new biomarker for CRC diagnosis.
In addition to being biomarkers of cancer, circRNAs are also involved in regulating the life activities of cancer cells. There is evidence that changes in circRNA expression have a wide range of effects on the biological characteristics of liver cancer, and oncogenic circRNA CDR1as promotes the growth of cancer cells and inhibits apoptosis by acting as a sponge of miR-7 (11). It has also been reported that circPVRL3 targets miR-203 and affects the ERK1/2/Slug/E-cadherin signaling pathway to inhibit the invasion of gastric cancer cells (24). Similarly, in the results of this assay, we found that knockdown of circ_0000799 inhibited the proliferation, viability, invasion, migration of CRC and EMT process. It is indicated that circ_0000799 may be involved in CRC occurrence and metastasis.
Many studies have explored the ways in which circRNA exerts biological functions, and the following four ways are the most frequently proposed: serving as a sponge of miRNA (25), transcription regulation and alternative splicing (26), interacting with RNA-binding proteins (27), and directly participating in translational regulation (28). Acting as a sponge of miRNA is the most well-studied mechanism of action of circRNAs. The miRNAs are a class of small non-coding RNAs of about 20–25 nt in length (29), which can prevent messenger RNA (mRNA) translation by complementary binding with mRNA 3'-UTR region (30). Cao et al. reported that miR-647 was significantly down-regulated in gastric cancer tissues and cell lines, and it was closely related to reduced tumor size and metastasis, down-regulated tumor proliferation, and the expression level of genes related to metastasis (31). Zhang et al. demonstrated that the level of miR-647 was significantly decreased in non-small cell lung cancer, and miR-647 could promote the therapeutic effectiveness of argon-helium cryoablation and inhibit cell proliferation through targeting TRAF2 via the NF-κB signaling pathway (32). These results show that miR-647 is a class of tumor suppressor gene that can inhibit the tumorigenic phenotype of cells. Our study showed that miR-647 was significantly down-regulated in CRC tissues and cells, showing a negative correlation with the expression of circ_0000799 and acting as a molecular sponge of miR-647. Thus, circ_0000799 exerts its biological function by affecting the expression level of miR-647.
There have been reports that circRNA acts as a sponge of miRNA and is involved in tumor growth regulation (11,24). This study reports for the first time that circ_0000799 promotes the growth of CRC cells and CRC metastasis by acting as a sponge of miR-647 through bioinformatics analysis, PCR validation, and dual-luciferase reporter assay.
Conclusions
In summary, circ_0000799 promotes cell proliferation, viability, invasion, and migration, and the EMT process by serving as a sponge of miR-647, thereby accelerating CRC progression. Therefore, circ_0000799 can function as a new biomarker in the diagnosis of CRC and has the potential to be a new therapeutic target for this disease. However, the function of circRNA is complex and this assay only validates circ_0000799 function at the cellular level. Further exploration is needed to provide a more comprehensive data base for the use of circ_0000799.
Acknowledgments
Funding: This work was supported by the Heilongjiang Natural Science Foundation (No. H2018017).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1176/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1176/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1176/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted according to the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Ethics Committee of The Fifth Affiliated Hospital of Harbin Medical University (No. KY-2022-019), and all of the patients provided their written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Kong W. Clinical and pathological characteristics of colorectal cancer patients in different age groups. Jinan: Shandong University, 2021.
- Winawer S, Fletcher R, Rex D, et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence. Gastroenterology 2003;124:544-60. [Crossref] [PubMed]
- Köhne CH, Karthaus M, Mineur L, et al. Impact of Primary Tumour Location and Early Tumour Shrinkage on Outcomes in Patients with RAS Wild-Type Metastatic Colorectal Cancer Following First-Line FOLFIRI Plus Panitumumab. Drugs R D 2019;19:267-75. [Crossref] [PubMed]
- Amersi F, Agustin M, Ko CY. Colorectal cancer: epidemiology, risk factors, and health services. Clin Colon Rectal Surg 2005;18:133-40. [Crossref] [PubMed]
- Morabito A, De Maio E, Di Maio M, et al. Tyrosine kinase inhibitors of vascular endothelial growth factor receptors in clinical trials: current status and future directions. Oncologist 2006;11:753-64. [Crossref] [PubMed]
- Mohammad N, Singh SV, Malvi P, et al. Strategy to enhance efficacy of doxorubicin in solid tumor cells by methyl-β-cyclodextrin: Involvement of p53 and Fas receptor ligand complex. Sci Rep 2015;5:11853. [Crossref] [PubMed]
- Jung WB, Shin JY, Suh BJ. The Short-term Outcome and Safety of Laparoscopic Colorectal Cancer Resection in Very Elderly Patients. Korean J Gastroenterol 2017;69:291-7. [Crossref] [PubMed]
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381-8. [Crossref] [PubMed]
- Shafabakhsh R, Mirhosseini N, Chaichian S, et al. Could circRNA be a new biomarker for pre-eclampsia? Mol Reprod Dev 2019;86:1773-80. [Crossref] [PubMed]
- Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 2018;9:417. [Crossref] [PubMed]
- Li XN, Wang ZJ, Ye CX, et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res 2018;37:325. [Crossref] [PubMed]
- Bi J, Liu H, Cai Z, et al. Circ-BPTF promotes bladder cancer progression and recurrence through the miR-31-5p/RAB27A axis. Aging (Albany NY) 2018;10:1964-76. [Crossref] [PubMed]
- Fu L, Chen Q, Yao T, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget 2017;8:43878-88. [Crossref] [PubMed]
- Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 2017;388:208-19. [Crossref] [PubMed]
- Guo JN, Li J, Zhu CL, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther 2016;9:7451-8. [Crossref] [PubMed]
- Zhang R, Xu J, Zhao J, et al. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci 2018;22:118-26. [PubMed]
- Yuan Y, Liu W, Zhang Y, et al. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun 2018;503:870-5. [Crossref] [PubMed]
- Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol 2015;8:16020-5. [PubMed]
- Qin XP, Zhao GP, Yang C, et al. Effect of circ_0000745 on the Proliferation and Metastasis of Colorectal Cancer Cells by Targeting miR-296-5p. Acta Medicinae Universitatis Scientiae et Technologiae Huazhong 2022;51:31-7.
- Zhang S, Chen Z, Sun J, et al. CircRNA hsa_circRNA_0000069 promotes the proliferation, migration and invasion of cervical cancer through miR-873-5p/TUSC3 axis. Cancer Cell Int 2020;20:287. [Crossref] [PubMed]
- Rong X, Gao W, Yang X, et al. Downregulation of hsa_circ_0007534 restricts the proliferation and invasion of cervical cancer through regulating miR-498/BMI-1 signaling. Life Sci 2019;235:116785. [Crossref] [PubMed]
- Li B, Li X. Overexpression of hsa_circ_0007534 predicts unfavorable prognosis for osteosarcoma and regulates cell growth and apoptosis by affecting AKT/GSK-3β signaling pathway. Biomed Pharmacother 2018;107:860-6. [Crossref] [PubMed]
- Zheng Y, Liu W, Guo L, et al. The expression level of miR-203 in patients with gastric cancer and its clinical significance. Pathol Res Pract 2017;213:1515-8. [Crossref] [PubMed]
- Li JQ, Yang J, Zhou P, et al. The biological functions and regulations of competing endogenous RNA. Yi Chuan 2015;37:756-64. [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- AbouHaidar MG, Venkataraman S, Golshani A, et al. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci U S A 2014;111:14542-7. [Crossref] [PubMed]
- Salmanidis M, Pillman K, Goodall G, et al. Direct transcriptional regulation by nuclear microRNAs. Int J Biochem Cell Biol 2014;54:304-11. [Crossref] [PubMed]
- Ma H, Wang P, Li Y, et al. Decreased expression of serum miR-647 is associated with poor prognosis in gastric cancer. Int J Clin Exp Pathol 2019;12:2552-8. [PubMed]
- Cao W, Wei W, Zhan Z, et al. Role of miR-647 in human gastric cancer suppression. Oncol Rep 2017;37:1401-11. [Crossref] [PubMed]
- Zhang YS, Chen T, Cai YJ, et al. MicroRNA-647 promotes the therapeutic effectiveness of argon-helium cryoablation and inhibits cell proliferation through targeting TRAF2 via the NF-κB signaling pathway in non-small cell lung cancer. Onco Targets Ther 2018;11:6777-84. [Crossref] [PubMed]
(English Language Editor: J. Jones)


