
LINC00178 promotes cell growth, migration, and invasion in colorectal cancer in vitro and in vivo
Introduction
Colorectal cancer (CRC) is a very common gastrointestinal malignant tumor for which the incidence is increasing annually and the age at diagnosis is becoming younger (1,2). At present, surgical resection is mainly used in clinical practice, supplemented by preoperative or postoperative chemotherapy. Although CRC diagnosis and treatment methods have made significant progress, due to the hidden onset of colon cancer, most patients have already reached the middle or late stage at the time of diagnosis. The shortage of sensitive analytical methods to diagnose CRC in the early stage leads to poor prognosis and around more than half of the patients with a continuous progression eventually die due to metastasis and recurrence (3,4). Molecularly targeted therapeutic drugs have recently played a significant role in the comprehensive treatment of patients with advanced CRC. Molecularly targeted therapeutic drugs with different targets have different effects, but the clinical application of molecularly targeted drugs is still unclear. There are many problems that need to be further clarified (5-7). For that reason, the discovery of molecular targets for early detection and novel targets for treatment, as well as the identification of molecular pathways impacting the growth of CRC, are crucial.
Any RNA molecule longer than 200 nucleotides is considered a long non-coding RNA (lncRNA), and these molecules play important roles in pathophysiology. They regulate gene expression at the epigenetic, transcriptional, and post-transcriptional stages (8,9). Studies have found that a diversity of lncRNAs are strangely expressed in a variety of species and play different biological roles. An example is lncRNAs with tumor-promoting functions, the overexpression of which promotes tumor growth and cancer cell proliferation and metastasis, or exerts tumor suppressor functions. The over expression of lncRNA can inhibit the progression, migration or transportation, and metastasis of tumor cells (10-12). In addition to a high prevalence of metastasis and recurrence, previous research has shown that lncRNAs play a crucial role in the development and spread of CRCs (13); CRC tissues showed markedly elevated expression of lncRNA and NEAT1 genes. Further, in vivo cum in vitro experiments have shown that overexpression of lncRNA, and NEAT1 genes promoted the progression and metastasis of CRCs by activating the Wnt signaling pathway (14). In addition, the lncRNA, and SNHG11 in peripheral blood serve as a unique biological marker for an early prognosis and timely diagnosis of CRC (15). Many lncRNAs, including LBX2-AS1, have been linked to increased tumor cell proliferation and resistance to the chemotherapy medication 5-fluorouracil in CRC (16).
New evidence suggests that the non-coding RNA LINC00178, commonly known as urothelial carcinoma associated 1 (UCA1), has a role in controlling tumor growth, apoptosis, and invasion (17,18). Thyroid cancer cells' proliferation and metastasis were discovered to be induced by LINC00178 via the elimination of miR-497-3p (19). However, the mechanism of LINC00178 in CRC is unclear. Therefore, in-depth research into the molecular level mechanisms of CRC cell invasion and metastasis, as well as the identification of potential biomarkers, are critical for disease diagnosis and treatment. We present the following article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-940/rc).
Methods
Clinical sample collection
This experiment included 45 patients with CRC in the Affiliated Hospital of Nanjing University of Chinese Medicine, from March to September 2021. All the patients were pathologically diagnosed with CRC; immune- and other related underlying diseases were excluded. No chemotherapy or other treatments were given before surgery. Cancer tissue and matched non-cancer colorectal tissue samples were obtained within half an hour of tumor excision and stored in liquid nitrogen. Each participant and each examining physician completed an informed consent form. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Affiliated Hospital of Nanjing University of Chinese Medicine (No. 2021NL-034-01).
Cell culture and transfection
This experiment used CRC cell lines (LoVo, SW620, SW480, HT-29, and HCT116), and normal or control human colorectal mucosal cells (FHC) were purchased from the Shanghai Institute of Biological Sciences (Shanghai, China). These cells were grown in a 5% CO2 incubator in a medium containing 15% fetal bovine serum (FBS) from either Roswell Park Memorial Institute (RPMI)-1640 or Dulbecco’s modified Eagle medium (DMEM; Gibco, Grand Island, NY, USA). Passaging was carried out at a ratio of 1:3. The CRC cells were selected in logarithmic phase and then inoculated into 6-well plates and cultured overnight. When the confluency of cells attached to the wall reached about 50%, the pcDNA-178 overexpression recombinant vector or empty vector plasmid was transfected with lentivirus, short hairpin (sh)RNA-Control, shRNA-1, and shRNA-2 were transferred to the cells. For specific steps, refer to the instructions of the transfection reagent. The relevant indicators were detected 48 hours after the cells were transfected.
Fluorescence quantitative polymerase chain reaction
The total RNA in the transfected CRC cells was isolated using the one-step TRIzol method, then the RNA was purified. Total RNA was then analyzed for concentration and purity using NanoDrop (Thermo Fisher, Waltham, MA, USA). Complementary DNA (cDNA) was prepared using a reverse transcription (RT) kit. We took 2 µL of the RT product as template, GAPDH as the internal reference, and used SYBR Green PCRMaster Mix kit (Thermo Fisher) for quantitative polymerase chain reaction (qPCR) detection. Each reaction system in the 20 µL vial included 2 µL of RT product, 10 µL of 2× SYBR Green dye, 0.8 µL of both primers (forward and reverse), and 6.4 µL of double distilled water (ddH2O). The 45 cycles of the reaction consisted of 95 ℃ for 5 minutes, 60 ℃ for 30 seconds, and at 72 ℃. for 10 seconds. After the reaction, the relative quantitative 2-ΔΔCt method was used for analysis.
Cell Counting Kit-8 assay
After transfection, CRC cells were added to a 96-well plate, 2×103 cells were allocated to each well, and 6 duplicate (2x) wells were set up per group and placed in a 37 ℃ incubator for different durations of time. The Cell Counting Kit-8 (CCK-8) mixture was poured into each vial, which was then normally incubated at 37 ℃ for 4 hours in order to carry out the detection at 72 hours. The optical density (OD) value of each vial was then determined at a wavelength (=450 nm) using a microplate reader.
Clonogenic assay
After transfection, the CRC cells were digested by trypsin and then centrifuged for 3 minutes. After that, the supernatant was discarded, and the pellets were washed thoroughly using 3 mL of sterile phosphate-buffered saline (PBS) solution. Next, centrifugation was conducted again, and repeated 3 times. The culture medium was then added and a single cell suspension was made to adjust cell concentration to 600 cells/mL, and the prepared single-cell suspension was evenly planted in a 6-well plate for colony formation experiments. After every 3rd day, the culture media was monitored and changed, colonies were fixed with 4% paraformaldehyde and stained by crystal violet for 15 min at room temperature and photographed by a camera.
Transwell migration assay
After adjusting to 1×105 cells/mL in serum-free DMEM, 600 µL of DMEM with 10% FBS was introduced to the transwell’s bottom chamber. The upper chamber contained 200 µL of test cell suspension. The whole chamber was cultured overnight in a conventional incubator, a cotton swab was used to clean the cells in the upper chamber, and cells adhering to the lower membrane surface of the transwell’s lower chamber were fixed with 4% para-formaldehyde solution for 15 minutes before being stained in 0.05% crystal violet dye for 40 minutes. The cells in transit were tracked and counted under a microscope.
Western blot detection
We digested, centrifuged, and collected the cells 48 hours after transfection, added cell lysate to resuspend the cells, centrifuged and collected the supernatant containing total protein, and used a bicinchoninic acid (BCA) kit to quantify the protein. A solution of 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel was prepared, 50 µg of protein was loaded, and then transferred to a polyvinylidene fluoride (PVDF) membrane after electrophoresis separation. The solution was blocked in 5% non-fat dry milk for 120 minutes at 25 ℃, and diluted with E-cadherin, N-cadherin, Vimentin, and GAPDH primary antibodies at 1:600 according to the antibody instruction manual. The corresponding primary antibodies were added and incubated for 24 hours at 4 ℃ on a shaker. The next day, after being rinsed 3 times in tris-buffered saline with Tween 20 (TBST) solution, the membrane had secondary antibodies diluted 1:3,000 added to it and incubated at 25 ℃ for 1 hour. Images were captured by exposing the membrane to a chromogenic solution, washing it again, and then exposing it to darkness in the dark room. Using GAPDH as the internal reference, Quantity One software (Bio Rad Laboratories, Hercules, CA, USA) was used to analyze the gray value of protein bands.
Tumorigenic experiment
Male BALB/c nu/nu mice (weight: 16–18 g, 4–5 weeks old) purchased from the Laboratory Animal Center of Shanghai, Academy of Science Chinese (Shanghai, China). The transfected CRC cells were re-suspended in serum-free media, the cellular density of suspension was adjusted to 1×107/mL, and 0.1 mL of the suspended tumor cell fluid was injected hypodermically in the right lower limb of the nude mice. After 10 days, a vernier calliper (0.01 mm) was used to measure the length (L) and breadth (W) of tumor-bearing tissues at the site of inoculation, and the tumor’s volume (V) was computed using the formula V= (L×W2)/2. A tumor’s volume was assessed once every 5 days. After 30 days, the naked mice were euthanized, their tumor-bearing tissues were harvested and photographed, and then the total messenger RNA (mRNA) in the tissues was extracted for subsequent detection and analysis.
The animal experiment was conducted in the Laboratory Animal Center of Nantong University. Animal experiments were approved by the Laboratory Animal Ethics Committee of Nantong University (No. S20210317-501), in compliance with national guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Statistical analysis
All measurement data obtained in this experiment are expressed as . The difference between the 2 samples was analyzed by the analysis of variance (ANOVA) variance SNK-q method, and P<0.05 was considered statistically significant.
Results
CRC tissues are characterized by high levels of LINC00178 expression
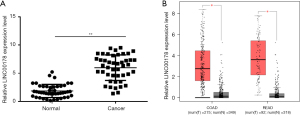
The expression of LINC00178 was first discovered by quantitative fluorescence (QF)-PCR in 45 early-stage colon cancer tissues and matched para neoplastic tissues to study its presence and relative abundance in tumors of the digestive tract. Thus, the results linked normal para cancerous tissues with LINC00178 and found that it was over expressed in cancer tissues (Figure 1A). Through the analysis of the transcriptome data related to colon cancer and rectal adenocarcinoma in The Cancer Genome Atlas (TCGA) database, in which, colon adenocarcinoma (COAD): 275 cases of cancer tissue and 349 cases of adjacent cancer; rectal adenocarcinoma (READ): 92 cases of cancer tissue and carcinoma 318 cases. It was shown that LINC00178 was differentially expressed in both COAD and READ (Figure 1B). The above results indicate that LINC00178 is over expressed in tissues.
Relationship between LINC00178 level and clinical case characteristics of colorectal cancer patients
To analyze the relationship between the expression level of LINC00178 and clinical characteristics of colorectal cancer patients. The results showed that LINC00178 expression was correlated with TNM stage and differentiation (all P<0.05) (Table 1).
Table 1
| Clinicopathological features | n | LINC00178 level | t | P |
|---|---|---|---|---|
| Gender | ||||
| Male | 25 | 5.67±1.73 | 0.7984 | 0.4290 |
| Female | 20 | 5.28±1.49 | ||
| Age (years) | ||||
| <65 | 11 | 5.64±1.64 | 0.6047 | 0.5486 |
| ≥65 | 34 | 6.01±1.80 | ||
| Tumor diameter | ||||
| <3 cm | 19 | 6.15±1.92 | 1.893 | 0.0651 |
| ≥3 cm | 26 | 5.21±1.45 | ||
| TNM stage | ||||
| I−II | 16 | 5.16±1.17 | 2.120 | 0.0398 |
| III−IV | 29 | 6.29±1.94 | ||
| Differentiation degree | ||||
| Poorly | 24 | 6.19±1.55 | 2.362 | 0.0228 |
| High | 21 | 5.18±1.28 |
Data are expressed as number and Mean ± SD. SD, standard deviation. TNM, tumor/lymph nodes/metastasis.
LINC00178 knockdown or overexpression influences CRC cells
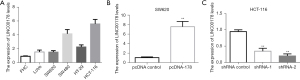
Additionally, to explore the role of LINC00178 in colorectal tumors, commonly used CRC cell-lines (LoVo, SW620, SW480, HT-29, and HCT116) and normal or control human colorectal mucosal cells (FHC) were first selected. The results of q-PCR compared human normal colorectal mucosal cells (FHC) with LINC00178 which was over expressed in CRC cell lines, with over expression in SW480 and HCT116, and relatively low expression in LoVo, SW620, and HT-29 (Figure 2A). According to the above results, the over expression vector (pcDNA-178) and blank control vector (pcDNA-control) of LINC00178 were synthesized and transfected into SW620 (with low expression of LINC00178). Compared with other groups, transfection of LINC00178 over expression vector (pcDNA-178) could significantly enhance the level of the expression level of LINC00178 in cells (Figure 2B). In addition, HCT-116 cells were transfected with 2 blank/control vector (shRNA-control) and LINC00178 knockdown vectors (shRNA-1 and shRNA-2). Transfection of LINC00178 knock down vectors (shRNA-1 and -2) dramatically decreased LINC00178 expression in cells than in the control group (Figure 2C).
CCK-8 measured the impact of LINC00178 overexpression and knockdown on the growth of CRC cells
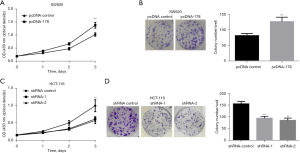
The results compared the blank control vector (pcDNA-control) group with transfection of the LINC00178 overexpression vector (pcDNA-178), which significantly promoted cellular proliferation (Figure 3A). In comparison with the control group, transfection of the LINC00178 overexpression vector (pcDNA-178) could significantly enhance the clonogenic ability of the cells (Figure 3B). Comparing the transfection of LINC00178 blank/control vector group (shRNA-control) and knockdown vectors (shRNA-1 and -2) showed that the latter could more effectively suppress cell growth (Figure 3C). Furthermore, the clonogenic experiment demonstrated a substantial reduction in clone formation capacity between the shRNA-control group and the transfection of LINC00178 knock down vectors (shRNA-1 and -2) (Figure 3D).
Knockdown or overexpression of LINC00178 can significantly affect CRC cell migration
Transwell assay was used to assess the effects of LINC00178 knockdown or overexpression level on the ability migration in colorectal malignant cells. This study compared SW680 cells transfected with the LINC00178 overexpression vector (pcDNA-178) to a batch of control cells transfected with a non-targeting vector (pcDNA-control). Which significantly promotes the ability of cells to migrate from the upper chamber of the transwell to the lower chamber. As a means of simulating the matrix environment in vivo, Matrigel was streaked over the top chamber of a Transwell, and the cells' capacity to invade was then measured. Invasion of SW680 cells from Matrigel through the top chamber of transwell and into the bottom chamber was much improved after transfection with LINC00178 overexpression vector (pcDNA-178) compared to the (pcDNA-control) group (Figure 4A). After LINC00178 was knocked down, the same approach was used to detect HCT-116 cell movement and invasion. The HCT-116 cells were transfected with a knockdown vector for LINC00178 and compared to a group transfected with a blank/control vector (shRNA-1 and shRNA-control). Cell migration and invasion from the upper transwell chamber to the lower chamber can be significantly inhibited (Figure 4A).
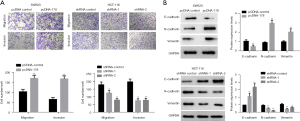
Western blot analysis revealed that the blank control vector (pcDNA-control) group surpassed the LINC00178 overexpression vector-transfected SW680 cells (pcDNA-178). E-cadherin expression was downregulated, but N-cadherin and vimentin levels were dramatically upregulated (Figure 4), through which cells underwent epithelial-mesenchymal transition (EMT) with favorable results. Transfection of LINC00178 knockdown vectors (shRNA-1 and -2) into HCT-116 cells led to a considerable up-regulation of E-cadherin expression compared to the control vector (shRNA-control) group, whereas it led to a significant down-regulation of vimentin expression. Vimentin and N-cadherin were significantly down-regulated (Figure 4B), inhibiting cells from undergoing EMT.
LINC00178 overexpression affects CRC cell growth in vivo
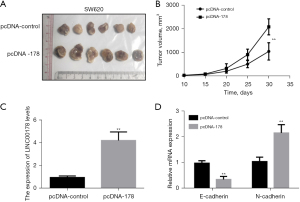
We investigated the impact of LINC00178 overexpression on CRC cell proliferation in vivo. First, nude mice were subcutaneously implanted with SW680 cells that had been transfected with an overexpression vector for LINC00178 (pcDNA-178). Mice were kept under specific pathogen-free conditions for 30 days before being slaughtered, at which point the tumors were removed and documented. The results compared the blank control vector (pcDNA-control) group with the tumor-bearing tissue of the LINC00178 overexpression vector (pcDNA-178) group was significantly increased (Figure 5A). Tumor-bearing tissues grew faster in the vector (pcDNA-178) group than in the blank/control vector (pcDNA-control) group (Figure 5B). The QF-PCR detected the expression levels of LINC00178 in tumor-bearing tissues and the results showed that those of the blank control vector (pcDNA-control) group compared with the in the tumor-bearing tissues of the LINC00178 overexpression vector (pcDNA-178) group was significantly increased (Figure 5C). In addition, QF-PCR detected the levels of expression in E-cadherin and N-cadherin inside tumor-bearing tissues. The expression levels of E-cadherin were significantly lowered, while the variable level of N-cadherin was significantly enhanced (Figure 5D).
Discussion
Accumulating evidence suggests that dysregulated lncRNA expression is linked with tumor initiation, proliferation, and poor prognosis, and can be used as novel biomarkers for early diagnosis of tumors (11,13,20). LncRNA has potential application value in the diagnosis, prognosis assessment and targeted therapy of CRC. LncRNA plays an important role in the occurrence and development of CRC by affecting key cancer signaling pathways, such as Wnt/β-catenin, PI3K/AKT, EGFR, NOTCH, m TOR and TP53 (21). Therefore, elucidating the function and regulatory mechanism of lncRNA in CRC has important potential to provide potential molecular targets for early correct diagnosis and targeted treatment of CRC. The results of this experiment revealed that LINC00178 is overexpressed in colorectal tumor tissues. LINC00178 expression is correlated with TNM stage and differentiation degree of colorectal cancer patients. The ability of CRC cells to proliferate and form clones is greatly aided by high levels of LINC00178 expression. Concurrently, the expression level of E-cadherin is dramatically lowered, and the number of cells that migrate and invade is significantly increased. Overexpression of N-cadherin and Vimentin regulates EMT pathways, which in turn affects the ability of CRC to invade and metastasize. Further, blocking LINC00178 expression reduced CRC cell growth in vivo.
Previous researches have shown that multiple genomic changes contribute to the development of CRC (22,23). One of these changes is the presence of lncRNA, which may regulate gene expression at the transcriptional, post-transcriptional, and epigenetic levels, thereby playing a role in tumor proliferation, invasion, metastasis, and related pathological processes. The CRC cells, for instance, may be pushed forward by lncRNA-cCSC1, which does so by silencing miR-124-3p and, in turn, increasing CD44 expression (24). In addition, lncRNA and CCAT2 are overexpressed in colorectal tumors, and promote the development and induce proliferation of tumor cells in order to inhibit apoptosis (25). The results highlight the possibility that lncRNAs play crucial regulatory roles in the development and metastasis of CRC cells. Results from this research demonstrated that LINC00178 was substantially expressed in CRC tissues, as shown by both clinical samples and TCGA data. Further, cellular functional experiments showed that high expression of LINC00178 induced cell proliferation while knockdown of LINC00178 thus inhibited cellular proliferation.
Tumor metastasis is the propagation of tumor cells from their primary site and the development of secondary tumors at distant sites. In most cases, metastatic CRC remains incurable and frequently leads to death in most patients (26,27). It has been demonstrated, for instance (28) that the lncRNA FAS1 regulates the miR-34b/SOX4 signaling axis, hence promoting CRC metastasis and invasion. The lymphatic spread of CRCs is aided by the lncRNA GAS5's high expression (29). Therefore, early identification and site-directed therapy of CRC depend on pinpointing the essential regulatory molecules that control invasion and metastasis.
In this work, we found that LINC00178 induces EMT process in CRC cells. Overexpressing LINC00178 increased the expression of N-cadherin and vimentin and decreased expression of E-cadherin, all of which facilitated cell invasion and migration. These results indicated that LINC00178 could promote metastasis of CRC cells through EMT pathway. Overexpression of LINC00178 greatly increased the growth of CRC cells in vivo tumorigenic experiments. However, it is still unclear how LINC00178 functions. Therefore, further experiments will continue to explore the localization of LINC00178 in the cytoplasm and nucleus, which is due to the different localization of lncRNA in cells and different mechanisms of its effect (30).
In conclusion, overexpression of LINC00178 is seen in CRC cells and tissues. Knockdown of LINC00178 has a variety of effects, whereas overexpression of LINC00178 has been shown to promote CRC cell growth, proliferation, clonal expansion, and invasion. The protein expression levels of E-cadherin, N-cadherin, and vimentin can all be regulated by the overexpression of LINC00178. Experiments using nude mice demonstrated that LINC00178 overexpression dramatically increased tumor cell proliferation in colorectal malignancies, indicating that this gene may represent a promising target for the treatment of this disease.
Acknowledgments
Funding: This study was supported by the Jiangsu Provincial Hospital of Traditional Chinese Medicine Project (NO. Y2020CX10).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-940/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-940/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-940/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Affiliated Hospital of Nanjing University of Chinese Medicine (No. 2021NL-034-01). Each participant and each examining physician completed an informed consent form. Animal experiments were approved by the Laboratory Animal Ethics Committee of Nantong University (No. S20210317-501), in compliance with national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li J, Ma X, Chakravarti D, et al. Genetic and biological hallmarks of colorectal cancer. Genes Dev
- 35:787-820.
- Kanth P, Inadomi JM. Screening and prevention of colorectal cancer. BMJ 2021;374:n1855. [Crossref] [PubMed]
- Fan A, Wang B, Wang X, et al. Immunotherapy in colorectal cancer: current achievements and future perspective. Int J Biol Sci 2021;17:3837-49. [Crossref] [PubMed]
- Li N, Lu B, Luo C, et al. Incidence, mortality, survival, risk factor and screening of colorectal cancer: A comparison among China, Europe, and northern America. Cancer Lett 2021;522:255-68. [Crossref] [PubMed]
- Shah SC, Itzkowitz SH. Colorectal Cancer in Inflammatory Bowel Disease: Mechanisms and Management. Gastroenterology 2022;162:715-730.e3. [Crossref] [PubMed]
- Baidoun F, Elshiwy K, Elkeraie Y, et al. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr Drug Targets 2021;22:998-1009. [Crossref] [PubMed]
- Piawah S, Venook AP. Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 2019;125:4139-47. [Crossref] [PubMed]
- Ali T, Grote P. Beyond the RNA-dependent function of LncRNA genes. Elife 2020;9:60583. [Crossref] [PubMed]
- Tan YT, Lin JF, Li T, et al. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond) 2021;41:109-20. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- McCabe EM, Rasmussen TP. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol 2021;75:38-48. [Crossref] [PubMed]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Wang L, Cho KB, Li Y, et al. Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer. Int J Mol Sci 2019;20:5758. [Crossref] [PubMed]
- Zhang M, Weng W, Zhang Q, et al. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol 2018;11:113. [Crossref] [PubMed]
- Xu W, Zhou G, Wang H, et al. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer 2020;146:2901-12. [Crossref] [PubMed]
- Ma YN, Hong YG, Yu GY, et al. LncRNA LBX2-AS1 promotes colorectal cancer progression and 5-fluorouracil resistance. Cancer Cell Int 2021;21:501. [Crossref] [PubMed]
- Yang A, Liu X, Liu P, et al. LncRNA UCA1 promotes development of gastric cancer via the miR-145/MYO6 axis. Cell Mol Biol Lett 2021;26:33. [Crossref] [PubMed]
- Liu T, Wang X, Zhai J, et al. Long Noncoding RNA UCA1 Facilitates Endometrial Cancer Development by Regulating KLF5 and RXFP1 Gene Expressions. Cancer Biother Radiopharm 2021;36:521-33. [Crossref] [PubMed]
- Gao H, Yang JY, Tong LX, et al. Long noncoding RNA UCA1 promotes proliferation and metastasis of thyroid cancer cells by sponging miR-497-3p. Eur Rev Med Pharmacol Sci 2020;24:7555. [PubMed]
- Shi SH, Jiang J, Sun L, et al. Dynamic Regulative Biomarker: Long Noncoding RNA (lncRNA) in Metastatic Breast Cancer. Clin Lab 2020;66. [Crossref] [PubMed]
- Galamb O, Barták BK, Kalmár A, et al. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J Gastroenterol 2019;25:5026-48. [Crossref] [PubMed]
- Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer 2020;19:167. [Crossref] [PubMed]
- Tang C, Liu J, Hu Q, et al. Metastatic colorectal cancer: Perspectives on long non-coding RNAs and promising therapeutics. Eur J Pharmacol 2021;908:174367. [Crossref] [PubMed]
- Zhang HR, Wu SY, Fu ZX. LncRNA-cCSC1 promotes cell proliferation of colorectal cancer through sponging miR-124-3p and upregulating CD44. Biochem Biophys Res Commun 2021;557:228-35. [Crossref] [PubMed]
- Gao P, Sun D, Guo H, et al. LncRNA CCAT2 promotes proliferation and suppresses apoptosis of colorectal cancer cells. J BUON 2020;25:1840-6. [PubMed]
- Cao H, Xu E, Liu H, et al. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol Res Pract 2015;211:557-69. [Crossref] [PubMed]
- Jin M, Frankel WL. Lymph Node Metastasis in Colorectal Cancer. Surg Oncol Clin N Am 2018;27:401-12. [Crossref] [PubMed]
- Deng H, Wang M, Xu Q, et al. ZFAS1 Promotes Colorectal Cancer Metastasis Through Modulating miR-34b/SOX4 Targeting. Cell Biochem Biophys 2021;79:387-96. [Crossref] [PubMed]
- Zheng Y, Song D, Xiao K, et al. LncRNA GAS5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget 2016;7:83727-34. [Crossref] [PubMed]
- Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol 2021;220:e202009045. [Crossref] [PubMed]
(English Language Editor: J. Jones)


