
c-MYC and HIF1α promoter G-quadruplexes dependent metabolic regulation mechanism of berberine in colon cancer
Introduction
The incidence and mortality of colon cancer are at the forefront of tumor diseases (1). It is worth noting that the incidence in young people under 50 years old is increasing significantly, especially for people aged 20 to 34 years old, and the reasons remain to be explored (2). Finding effective prevention and treatment methods of colon cancer is still a challenge. At present, the conventional treatment of colon cancer mainly involves surgery, chemotherapy, radiotherapy, immunotherapy, targeted therapy, and traditional Chinese medicine, among other methods. However, there is still a lack of accurate treatment methods, and drug research and development based on effective targets is an urgent need.
Bioinformatics studies have shown that gene biomarkers of colon cancer are closely related to prognosis (3-5). In fact, one gene may regulate a variety of biological processes, and numerous different genes constitute the molecular network regulating biological activities. Intervening in the expression of key genes may affect multiple biological processes of the tumor at the same time. In addition to excessive cell proliferation, abnormal apoptosis, migration, and invasion, metabolic changes are also important characteristics of tumors. Genes involved in the regulation of tumor biological effects such as cell proliferation, apoptosis, migration, and invasion also play an important role in the regulation of tumor metabolism (6). For instance, tumor suppressor p53, proto-oncogene MYC (mainly c-MYC), and hypoxia inducible factor 1 (HIF1) may participate in the tricarboxylic acid (TCA) cycle and other processes to varying degrees. p53 can combine with glucose-6-phosphate dehydrogenase (G6PD) and inhibit its activity, so as to block the pentose phosphate pathway and inhibit tumor processes (7). c-MYC and HIF1 are more closely related to tumor metabolic reprogramming. c-MYC can regulate aerobic glycolysis and oxidative phosphorylation, promote the production of adenosine triphosphate (ATP), and accelerate tumorigenesis and metastasis. HIF1α can accelerate tumor metastasis by promoting aerobic glycolysis (8). Exploring the small molecules that can affect the biological activities of such key genes regulating cell proliferation, apoptosis, migration, invasion, and metabolism in cancer processes may provide a new strategy for the development of anticancer drugs.
The G-quadruplex, a special secondary structure of nucleic acids formed by guanine-rich sequences, widely exists in telomeres, oncogene promoters, microsatellite fragments, and other regions in the human body (9-12). With the development of genomics, more and more G-quadruplex structures have been found in organisms (13-15). G-quadruplex plays the role of a molecular switch regulating gene transcription in biological processes, and can inhibit or promote gene transcription, then intervene in the translation of gene expression and further affect related signaling pathways (11,14). For example, maintaining the structural stability of the G-quadruplex located in the telomere or promoter regions of oncogenes such as KRAS and c-MYC and other genes such as CLIC4 is conducive to inhibiting gene transcription and delaying tumor processes (16-18), while maintaining the structural stability of the G-quadruplex in some microRNAs is useful for promoting the transcription of target genes (11). Therefore, G-quadruplex structures were considered to be effective targets for the treatment of cancer, and many new drugs based on molecular recognition of the G-quadruplex have been developed (19). In that case, c-MYC and HIF1α G-quadruplexes may be potential targets for metabolic regulation in cancer. However, there are still some unknown problems in this field, including whether the interaction properties between small molecule drugs and G-quadruplexes of various genes are the same, and more importantly, whether small molecule drugs exert anticancer effects by interacting with the G-quadruplex of different genes simultaneously. Further exploration needs to be performed to clarify these problems.
Berberine is an important natural product, the drug form of which is called hydrochloride berberine. In the pharmaceutical industry, it can be extracted from a variety of medicinal plants or obtained by chemical synthesis. Berberine is used clinically as a drug for the treatment of intestinal infection. In addition, berberine also has pharmacological effects such as hypoglycemic, lipid-lowering, anticancer, and anti-inflammatory effects, among others. Basic studies have shown that berberine has inhibitory effects on a variety of tumors, especially colorectal cancer (20-23). The prospective, multicenter, double-blind, randomized, placebo-controlled clinical study confirmed that berberine could reduce the recurrence of colorectal adenoma (24,25). So far, studies have shown that berberine can play an anti-colon cancer role through AMPK, β-catenin, and other signaling pathways, but other mechanisms such as the metabolic regulation mechanism of berberine in colon cancer is not yet clear (23,26). Previous studies confirmed that there was also a parallel G-quadruplex structure in the promoter region of HIF1α (27), as well as KRAS and c-MYC (17,18), and berberine could bind with KRAS and c-MYC promoter G-quadruplexes (28,29). Nevertheless, whether berberine can combine with HIF1α G-quadruplex and further regulate metabolism in colon cancer based on G-quadruplexes in promoter regions of c-MYC and HIF1α remained to be explored. In this study, we aimed to clarify the following questions: first, whether the interacting ability of berberine with HIF1α and c-MYC G-quadruplexes is similar; second, what is the influence of berberine on the biological effects related to c-MYC and HIF1α in colon cancer cells; third, whether berberine regulates metabolism based on interacting with c-MYC and HIF1α G-quadruplexes in colon cancer. In that way, we adopted a variety of spectroscopy methods to study the binding mode of berberine with c-MYC and HIF1α G-quadruplexes, investigated the effects of berberine on c-MYC and HIF1α related biological effects by MTT, flow cytometry, quantitative real-time polymerase chain reaction and western blot, and finally explored the effects of berberine on TCA cycle and aerobic glycolysis via transcriptome sequencing in cell and central carbon metabolism analysis in vivo. We present the following article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-389/rc).
Methods
Materials
Berberine Chloride Hydrate was purchased from TCI development Co., Ltd (Tokyo, Japan). G-quadruplex sequences were synthesized by Sangon Biotech (Shanghai) Co., Ltd (Shanghai, China). Colonic cancer cell lines HCT116 and CT26 cells were ordered from Nanjing COBIOER Biotechnology Co. Ltd (Nanjing, China); phosphate belanced solution (PBS), trypsin and Trizol were ordered from the Jiangsu KeyGEN BioTECH Co. Ltd (Nanjing, China); Roswell Park Memorial Institute-1640 (RPMI-1640) basic culture medium and fetal bovine serum (FBS) were ordered from Corning Co. Ltd (Carlsbad, CA, USA). Mass spectrometry (MS) grade methanol, formic acid and ammonium acetate were from Fisher Scientific Technology Co. Ltd. (Waltham, MA, USA). Ultra-pure water was produced by a Milli-Q Ultra-pure water system (Millipore, Billerica, MA, USA).
Preparation of the working solution for spectroscopy analysis
The c-MYC and HIF1α G-quadruplex sequences were dissolved in Tris-HCl buffer solution (pH 7.4, 100 mM KCl) to make the concentration 100 µM, heated at 95 ℃ for 5 min, cooled naturally to room temperature, and then stored at 4 ℃ for use. Berberine hydrochloride was prepared into a 1-mM stock solution.
Ultraviolet and visible (UV-Vis) absorption spectroscopy
The concentration of berberine hydrochloride was fixed at 10 µM in Tris-HCl buffer solution (pH 7.4, 100 mM KCl). Different concentrations of c-MYC or HIF1α G-quadruplexes were mixed with berberine hydrochloride separately for 30 s, then the absorption spectra were measured by the U-3900H Spectrophotometer (Hitachi Limited, Japan) with the slit width and sampling interval set at 1 nm, until the absorbance no longer changed at room temperature. Every experiment was performed 3 times.
Fluorescence spectroscopy
Berberine hydrochloride solution with a concentration of 2.5 µM in Tris-HCl buffer (pH 7.4, 100 mM KCl) was mixed with different concentrations of G-quadruplex DNA until the fluorescence emission intensity no longer changed. After mixing and standing for 30 s, the fluorescence emission spectra of berberine were measured by the F-7000 FL Spectrophotometer (Hitachi Limited, Japan) at 293 K. For the equimolar job plots, the molar ratio of berberine hydrochloride to the G-quadruplex was adjusted from 0:10 to 10:0, and the fluorescence emission spectra were measured as mentioned at 293 K. The excitation wavelength was 345 nm, and the excitation and emission slits were both 5 nm. The fluorescence emission spectra of berberine hydrochloride binding with 10 different concentrations of c-MYC and HIF1α G-quadruplexes were measured at 313 K. The variation degree of fluorescence emission intensity of berberine binding with c-MYC or HIF1α G-quadruplexes at different temperatures was expressed as (F-F0)/F0 (F0 stood for the initial fluorescence intensity of berberine, while F stood for the intensity after binding with G-quadruplexes). Every experiment was performed 3 times.
Circular dichroism (CD) spectroscopy
The CD spectra of 10 µM c-MYC or HIF1α G-quadruplexes mixed with different concentrations of berberine hydrochloride were obtained by the Chirascan spectrometer (Applied Photophysics Ltd., UK) at room temperature. The recorded wavelength range was 220 to 340 nm and the bandwidth was 1 nm. The baseline was corrected to eliminate the interference signal of Tris-HCl buffer (pH 7.4, 100 mM KCl). For the CD melting curve experiments, the molar ellipticity at 263 nm of 10 µM G-quadruplexes before and after mixing with 100 µM berberine hydrochloride was monitored during the temperature of control unit rised from 35 to 105 ℃. The heating rate was 2 ℃/min and the data were acquired at intervals of 0.5 ℃. The temperature of the tangent point of the melting curve was defined as Tm. Every experiment was performed 3 times.
Cell culture
HCT116 cells and CT26 cells were cultivated in RPMI-1640 culture medium supplemented with 10% heat-inactivated FBS and 1% penicillin and streptomycin, under the condition of 37 ℃ and 5% CO2 in an incubator. Subculture was carried out when the cells grew to 90%.
Cell proliferation and apoptosis assays
HCT116 cells were treated with or without different concentrations of berberine hydrochloride for 24 h for proliferation and apoptosis assays. Cell proliferation was measured by MTT assay following the reported protocols. Cell apoptosis was determined by FACS Calibur (Becton, Dickinson and Company, USA) flow cytometry using an Annexin V-APC/7-AAD apoptosis detection kit (#KGA1024, Kaiji, Nanjing, China) according to the instructions of the manufacturer.
Quantitative real-time polymerase chain reaction (qRT-PCR)
HCT116 cells were treated with 50 µM berberine hydrochloride for 24 h, and the cells without drug intervention were used as controls. The Trizol total RNA extraction kit was used to extract the total RNA of cell samples. The primers used are listed in Table 1. The one step TB green reverse transcription kit and realtime PCR Master Mix (SYBR Green) were used for amplification. The relative expression of mRNA was analyzed by the 2-ΔΔCT method.
Table 1
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| c-MYC | CTGCGACGAGGAGGAGGACT | GGCAGCAGCTCGAATTTCTTT |
| HIF1α | CGGCGCGAACGACAAGAAA | AAGTGGCAACTGATGAGCAAG |
| GAPDH | GACGGCCGCATCTTCTTGT | CACACCGACCTTCACCATTTT |
HIF1α, hypoxia inducible factor 1-alpha; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Protein sample preparation and western blot
HCT116 cells were treated with 50 µM berberine hydrochloride for 24 h, while those cultured in normal medium were used as controls. The whole protein extraction kit was used to extract the total protein of the cell samples, and the BCA protein content detection kit was used to determine the protein concentration of each sample according to the instructions of the manufacturer. Western blot analysis was performed as described previously (30). The imaging was performed using the G:BOXChemiXR5 (Syngene, UK) gel imaging system. Gel-Pro32 software was used to analyze the band intensity. The antibodies used were as follows: c-MYC polyclonal antibody (#10828-1-AP, Proteintech, USA), mouse monoclonal to HIF1α (#ab1, Abcam, UK), rabbit anti-GAPDH (#KGAA002, KeyGEN BioTECH, China), goat-anti-rabbit IgG-HRP (#KGAA35, KeyGEN BioTECH, China), and goat-anti-mouse IgG-HRP (#KGAA37, KeyGEN BioTECH, China).
Transcriptome sequencing and bioinformatics analysis
HCT116 cells were treated with 50 µM berberine hydrochloride for 24 h. Then, the berberine intervention group and blank control group of cells were collected, and total RNA was extracted with Trizol. The integrity, purity, concentration, and RNA integrity number were analyzed successively. The cDNA library was generated and further purified, and the insert size and the effective concentration of the library were detected to ensure the library quality. Finally, the cDNA library was sequenced using the NovaSeq 6000 system (Illumina, USA). Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) was used to estimate gene expression levels. The screening criteria of differential genes were |log2FC| >1 and P<0.05 between the two groups. The metabolism gene set was obtained from the Molecular Signatures Database (MSigDB). Metabolic differentially expressed genes by RNA sequencing were extracted to perform enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways through R software.
Animal experiments
The animal experiment was approved by the Scientific Research Ethics Committee of Beijing Shijitan Hospital, Capital Medical University (Project No. 2018 scientific research ethics review No. 70). The animal experiment protocol followed the guidelines of China legislations on the ethical use and care of laboratory animals. A protocol was prepared before the study without registration.
Healthy male Balb/c mice (6 weeks old, 17–19 g) were obtained from Beijing Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China). All mice had free access to standard rat chow and water and were raised in a specific pathogen free (SPF) grade standard environment (50%±10% relative humidity, 12:12 h light-dark cycle, 22±2 ℃). After 1 week, CT26 cells (3×106 cells/100 µL) were injected into the right flank of 16 Balb/c nude mice subcutaneously. On the second day of the inoculation, the mice were randomly divided into two groups (eight in each group) by the random number table: one group received berberine hydrochloride diluted in 0.5% carboxymethyl cellulose sodium (CMC-Na) solvent with daily oral gavage of 50 mg·kg−1, and the other group received solvent as a control. The tumor size was measured every two days and assessed according to the formula: 0.5 × tumor length × tumor width2. After 14 days, the mice with the maximum and minimum tumor volume were excluded, and 6 mice were retained in each group for tumor biology study and central carbon metabolism analysis. The mice were anesthetized with CO2, the tumors were taken out and weighed, then one part was fixed with formalin and the other parts were kept at −80 ℃. In order to minimize errors, the operation was carried out by the same researcher and the mice were treated in the same order. Researchers were aware of the group allocation at the different stages of the experiment.
Hematoxylin and eosin (H&E) and immunofluorescence (IF) analysis
The detailed procedures were followed as the literature reports (31,32). In brief, tumor tissues fixed in formalin were dehydrated in alcohol, permeated in xylene, embedded in paraffin, cut into 4 µm thick slices, collected on slides in 40 ℃ water, and dried for use. For H&E staining, the slices were dewaxed, covered with water, stained with H&E, dehydrated, infiltrated with xylene, and sealed with neutral gum. Whole slide imaging was performed by the Pannoramic MIDI digital slice scanner (3DHISTECH, Hungary). For IF staining, the slices were dewaxed, placed in EDTA buffer for antigen retrieval, blocked with 5% goat serum at room temperature for 30 min, incubated with the primary antibody (1:250) at 4 ℃ overnight, and incubated with secondary antibodies of the corresponding species for 1h in the dark at room temperature. Sections were then stained with DAPI and sealed with the sealing agent. Images were taken using the Eclipse Ti-SR fluorescence inverted microscope (Nikon, Japan). The primary antibodies were Myc-Tag (9B11) mouse mAb (#2276, CST, USA), rabbit monoclonal to HIF1α (#ab179483, Abcam, UK), goat anti-rabbit IgG H&L (FITC) (#ab6717, Abcam, UK), and goat anti-mouse IgG H&L (Cy3®) preadsorbed (#ab97035, Abcam, UK).
High performance ion chromatography-mass spectrum/mass spectrum (HPIC-MS/MS) analysis of central carbon metabolism
An aliquot of each of the standard substance stock solutions was mixed to form the working standard solution. A series of calibration standard solutions were then prepared by stepwise dilution of this mixed standard solution. The weighed tissue samples were mixed with 500 µL precooled MeOH/H2O (3/1, v/v), then vortexed, homogenized, and sonicated in an ice-water bath and centrifuged at 13,800 g at 4 ℃ for 15 min. The supernatants were dried by spinning, and the residue was reconstituted in water and filtered through a 0.22-mm filter membrane for use.
The HPIC separation was carried out using a Thermo Scientific Dionex ICS-6000 HPIC System (Thermo Scientific, USA) equipped with Dionex IonPac AG11-HC (2 mm × 50 mm) and AS11-HC (2 mm × 250 mm) columns. The mobile phase A was 100 mM NaOH in water and the mobile phase D was ultrapure water. Another solvent (2 mM acetic acid in methanol) was added after the pump before entering the electrospray ionization (ESI) source (flow rate of 0.15 mL/min). The column temperature was set at 30 ℃. The injection volume was 5 µL. The AB Sciex QTRAP 6500+ triple quadrupole mass spectrometer (AB Sciex), equipped with an ESI interface, was applied and multiple reaction monitoring (MRM) mode was adopted for assay development. Typical ion source parameters were: IonSpray voltage =−4,500 V, temperature =450 ℃, ion source gas 1 =45 psi, ion source gas 2 =45 psi, curtain gas = 30 psi. AB Sciex Analyst Work Station Software 1.6.3, MultiQuant 3.0.3, and Chromeleon7 were employed for MRM data acquisition and processing. The signal-to-noise ratios (S/N) of 3 and 10 were used to determine the lower limits of detection (LLODs) and lower limits of quantitation (LLOQs), respectively. Methodological investigations such as precision and accuracy were implemented to ensure the reliability of test results. The metabolites with variable importance in the projection (VIP) >1 and P<0.1 were defined as differential metabolites.
Enrichment of metabolic pathways
The differential metabolites were searched by pathway analysis on the MetaboAnalyst 5.0 website (https://www.metaboanalyst.ca/home.xhtml) to obtain metabolic pathways.
Statistical analysis
Tumor volume, tumor mass and metabolite concentration were expressed in the form of (). The differences between the model and treatment groups were analyzed through the t-test by SPSS v19.0 (IBM, Armonk, NY, USA), and P<0.05 indicated statistical significance.
Results
Interaction characteristics revealed by UV-Vis absorption spectra
In order to explore the interaction characteristics of berberine with c-MYC and HIF1α promoter G-quadruplexes, the UV-Vis absorption spectra of berberine before and after interaction were acquired at room temperature. As shown in Figure 1A, after binding with increasing concentrations of c-MYC G-quadruplex, the maximum absorbance of berberine at 345 nm gradually decreased (called hypochromicity) with the red shift of the absorption wavelength. The detailed parameters are shown in Table 2, and the hypochromicity and red shift effect of the absorbance at 345 nm were 24.3% and 6 nm, respectively. In addition, there were some isosbestic points, implying the formation of combination equilibrium. For berberine binding with HIF1α G-quadruplex, it can be seen that the hypochromicity and red shift effect of the maximum absorbance at 345 nm were 34.3% and 7 nm, respectively. The hypochromicity was more obvious than that of berberine binding with c-MYC G-quadruplex, and isosbestic points could also be observed (see Figure 1B and Table 2). The UV-Vis absorption spectra characteristics indicated that there were π-π stacking interactions between berberine and c-MYC and HIF1α G-quadruplexes.
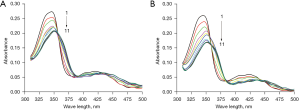
Table 2
| Gene | λfree/nm | λbound/nm | ∆λ/nm | Hypochromicity % |
|---|---|---|---|---|
| c-MYC | 345 | 351 | 6 | 24.3 |
| HIF1α | 345 | 352 | 7 | 34.3 |
HIF1α, hypoxia inducible factor 1-alpha.
Binding mode revealed by fluorescence emission spectra
Fluorescence emission spectroscopy is an effective means to study the interaction mode between small molecules and biological macromolecules. The fluorescence emission of berberine was relatively weak. When mixed with increasing concentrations of c-MYC G-quadruplex, the fluorescence emission of berberine increased significantly in turn at 293 K. When the fluorescence intensity no longer changed, the concentration of c-MYC G-quadruplex was 8 µM, as shown in Figure 2A. At the same concentration, the fluorescence emission of berberine combined with HIF1α G-quadruplex enhanced more significantly, and the G-quadruplex concentration was 7.2 µM when the fluorescence intensity no longer changed, as shown in Figure 2B. The increasing fluorescence emission also indicated that there were π-π stacking interactions between berberine and c-MYC and HIF1α G-quadruplexes.
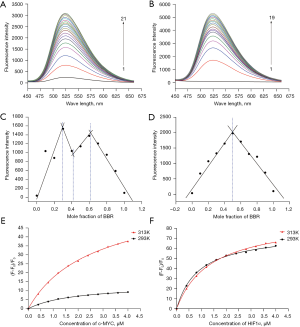
The stoichiometric ratio of berberine to c-MYC or HIF1α G-quadruplex was evaluated by equimolar job plots. It could be inferred that there were 3 stoichiometric ratios, 1:1, 1:3, and 3:1, between berberine and c-MYC G-quadruplex (Figure 2C). However, for berberine binding with HIF1α G-quadruplex, there was only one stoichiometric ratio of 1:1 (Figure 2D).
At 313 K, the fluorescence emission of berberine also enhanced after it bound with c-MYC or HIF1α G-quadruplex, but the intensity was weaker than that at the same concentration at 293 K (see Figure S1). It can be concluded that following the change of temperature, the variation degree of the fluorescence emission of berberine binding with c-MYC G-quadruplex was more significant than that of berberine binding with HIF1α G-quadruplex (Figure 2E,2F). Therefore, temperature has a greater effect on the combination of berberine with c-MYC G-quadruplex.
Berberine maintained the parallel configuration and increased the thermal stability of c-MYC and HIF1α G-quadruplexes
The effects of berberine on the configuration of c-MYC and HIF1α G-quadruplexes were studied by CD spectroscopy at room temperature. As depicted in Figure 3A, the c-MYC G-quadruplex sequence presented a positive peak at 263 nm and a negative peak at 242 nm, consistent with the characteristics of the parallel conformation. With the increasing concentration of berberine, the positive peak intensity decreased, but the parallel conformation did not change, indicating that berberine bound with c-MYC G-quadruplex and kept its parallel configuration. In Figure 3B, it was revealed that the HIF1α G-quadruplex sequence also presented a parallel conformation, and the effect of berberine on HIF1α G-quadruplex was similar to that on c-MYC G-quadruplex.
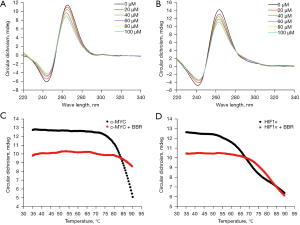
The effects of berberine on the thermal stability of c-MYC and HIF1α G-quadruplexes were explored by melting curve experiments. The Tm of c-MYC G-quadruplex was approximately 84 ℃, and increased to more than 90 ℃ after binding with berberine (Figure 3C). Meanwhile, the Tm of HIF1α G-quadruplex was approximately 70 ℃, and increased to approximately 80 ℃ after binding with berberine (Figure 3D). Accordingly, c-MYC G-quadruplex was more stable than HIF1α G-quadruplex, and berberine increased the thermal stability of both of them.
Berberine affected the proliferation and apoptosis of colon cancer cells
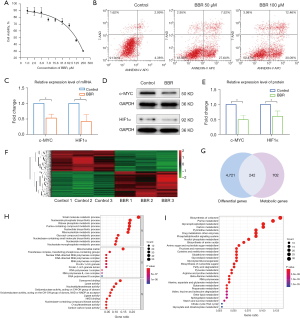
The effects of berberine on the proliferation and apoptosis of HCT116 cells were investigated. MTT assays showed that with the increasing concentration of berberine, the viability of HCT116 cells decreased gradually (Figure 4A). Flow cytometry assays indicated that berberine could promote the apoptosis of HCT116 cells in a dose-dependent manner (Figure 4B).
Berberine inhibited mRNA and protein expression of c-MYC and HIF1α in colon cancer cells
The effects of berberine on the mRNA and protein expression of c-MYC and HIF1α in HCT116 cells were studied by qRT-PCR and western blot analyses. The results demonstrated that berberine could inhibit the mRNA expression of c-MYC and HIF1α (Figure 4C) as well as the protein expression of c-MYC and HIF1α (see Figure 4D,4E).
Berberine may regulate the metabolism of colon cancer cells
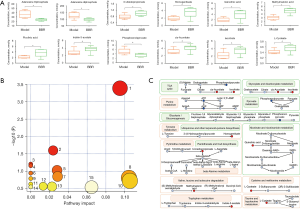
Transcriptome sequencing of HCT116 cells showed that there were 4,963 differentially expressed genes between the control group and berberine intervention group. Compared with the control group, 2,272 genes were downregulated and 2,691 genes were upregulated in the berberine intervention group, as illustrated by the heatmap in Figure 4F. In order to explore the effect of berberine on the metabolism of colon cancer cells, differential genes in the transcriptome were intersected with metabolic genes, and 242 differential metabolic regulatory genes were obtained (see Figure 4G). On this basis, GO and KEGG enrichment analyses were carried out. For GO enrichment, it was found that these differential metabolic regulatory genes were involved in biological processes (BPs) such as small molecule catabolic process, nucleoside phosphate biosynthetic process, and ribose phosphate metabolic process. They were also involved in the molecular functions (MFs) of coenzyme binding, lyase activity, and nucleotidyltransferase activity, among others, through the mitochondrial matrix, transferase complex, transferring phosphorus-containing groups, nuclear DNA-directed RNA polymerase complex, and other cellular components (CCs) (see Figure 4H). As for KEGG analysis, the top 30 metabolic pathways according to the arrangement of P value from small to large are shown in Figure 4I, which mainly contained pathways associated with central carbon metabolism, lipid metabolism, and amino acid metabolism, including biosynthesis of cofactors, purine metabolism, glycerophospholipid metabolism, carbon metabolism, pyrimidine metabolism, biosynthesis of amino acids, and so on.
Berberine delayed tumor progression in a colon cancer mouse model
Colon cancer cells were implanted into male Balb/c mice. On the next day, vehicle and berberine were administered by gavage to investigate the effect of berberine on tumor progression. It could be seen that at first, berberine had no significant effect on tumor growth. However, 14 days after administration, there was a significant difference in tumor size between the model group and berberine intervention group (see Figure 5A). The tumor mass in the drug intervention group was less than that in the model group, indicating that berberine could inhibit tumor growth (see Figure 5B).
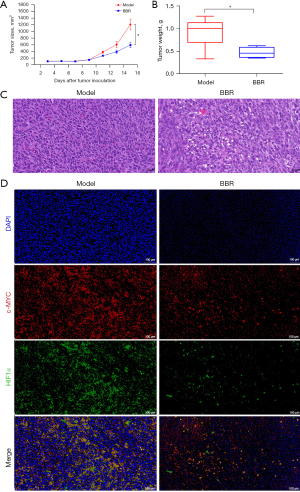
As shown in Figure 5C, the results of HE staining showed that the tumor cells in the model group were closely arranged, the proportion of nucleus to cytoplasm was large, and the nucleus was deeply stained. In comparison, in the berberine intervention group, the tumor cells were slightly sparse, the ratio of nucleus to cytoplasm decreased, the nuclear staining was slightly shallow, and fragmentation of the nucleus was obvious, indicating that berberine induced the damage of tumor cells.
The protein expression levels of c-MYC and HIF1α in the tumor tissues of colon cancer mice were detected by IF, as shown in Figure 5D. It was found that c-MYC and HIF1α were highly expressed in tumor tissues, but after being treated with berberine, their expression decreased to varying degrees, implying that berberine inhibited the protein expression of c-MYC and HIF1α.
Berberine regulated central carbon metabolism in the colon cancer mouse model
In order to evaluate the effect of berberine on metabolism associated with c-MYC, HIF1α, and nucleic acid, 50 metabolites associated with central carbon metabolism processes in tumor tissues were quantitatively detected by HPIC-MS/MS. It was found that between the model group and berberine intervention group, there were 12 differential metabolites with statistically significant differences (see Figure 6A, Table S1). The contents of ATP, adenosine diphosphate (ADP), and phosphoenolpyruvate decreased in the berberine intervention group compared with the model group, while the contents of 3-ureidopropionate, homogentisate, quinolinic acid, methylmalonic acid, picolinic acid, indole-3-acetate, cis-aconitate, isocitrate, and L-cysteate increased in the berberine intervention group compared with the model group. All the values of VIP for the 12 metabolites were >1.
The metabolic pathways involved in the differential metabolites were enriched through MetaboAnalyst 5.0. A total of 15 pathways were identified as shown in Figure 6B, and the specific information of the enriched pathways are shown in Table S2. The differential metabolites cis-aconitate, isocitrate, and phosphoenolpyruvate belonged to the citrate cycle (TCA cycle); phosphoenolpyruvate also belonged to pyruvate metabolism and glycolysis/gluconeogenesis; ATP and ADP belonged to purine metabolism; homogentisate belonged to tyrosine metabolism, ubiquinone, and other terpenoid-quinone biosynthesis; quinolinic acid belonged to nicotinate and nicotinamide metabolism; 3-ureidopropionate belonged to pantothenate and CoA biosynthesis, pyrimidine metabolism, and beta-alanine metabolism; methylmalonic acid belonged to valine, leucine, and isoleucine degradation; L-cysteate belonged to cysteine and methionine metabolism and taurine and hypotaurine metabolism; and indole-3-acetate belonged to tryptophan metabolism. Except for glyoxylate and dicarboxylate metabolism pathway, the above 14 pathways were found to be involved in the metabolic regulation of berberine in colon cancer revealed by central carbon metabolism research. Most animal and human cells do not have glyoxylic acid circulators, and acetyl CoA is unable to be converted into sugar, so that glyoxylate and dicarboxylate metabolism pathway mainly exists in plants, some microorganisms, and some invertebrate cells, and are therefore not discussed here. The metabolic transformation pathways of the differential metabolites are illustrated in Figure 6C. The metabolites phosphoenolpyruvate, homogentisate, 3-ureidopropionate, and L-cysteate participated in multiple metabolic pathways simultaneously, indicating that metabolic regulation is a complex network process.
The metabolic pathways enriched by central carbon metabolism detection were integrated with the metabolic pathways obtained by transcriptome sequencing of HCT116 cells. The intersection indicated that there were 13 common metabolic pathways, as shown in Figure S2 and Table S3.
Discussion
The G-quadruplex structure was first reported in 1962 (10). Four guanine molecules form G-tetrads through Hoogsteen hydrogen bonds, more than 3 G-tetrads form G-quadruplexes, and the central monovalent cation is the necessary factor to stabilize its structure. According to the spatial arrangement of bases, G-quadruplexes composed of different bases may have different configurations. Much experimental data have shown that there are 3 basic conformations: parallel conformation, antiparallel conformation, and hybrid conformation. The intramolecular and intermolecular G-quadruplexes can be formed based on a different number of molecules. The characteristics of the G-quadruplex structure determine its ability to interact with small molecule drugs. Folding into a G-quadruplex is not conducive to the combination of gene and transcription factor, so gene transcription can be inhibited in this situation. Instead, unfolding is conducive to the combination of gene and transcription factor. Therefore, the G-quadruplex is known as the molecular switch regulating gene transcription.
The occurrence of tumors is closely related to the abnormal expression of proto-oncogenes, among which the c-MYC gene is overexpressed in 70% of cancer cells. As a key node of the downstream signaling network, c-MYC is often activated in human solid tumors and is involved in the regulation of tumor biological processes such as cell proliferation and apoptosis. Therefore, it is an ideal target for cancer treatment. In addition to proto-oncogenes such as c-MYC, the role of HIF1α in the tumor microenvironment should not be ignored. Blocking HIF1α-related pathways can effectively inhibit tumor metastasis. Furthermore, it is known that cancer is considered a metabolic disease, including colon cancer (33). Tumor metabolic reprogramming is an important metabolic feature of cancer. When canceration occurs, tumor cells can obtain energy and proliferate rapidly through aerobic glycolysis (also named Warburg effect) and oxidative phosphorylation, which are 2 forms of tumor metabolic reprogramming (34). Therefore, metabolic regulation, especially that upon tumor metabolic reprogramming, may be an effective target for the prevention and treatment of colon cancer. Both c-MYC and HIF1α are associated with tumor metabolism, as they can regulate glucose transport, the TCA cycle, glycolysis, and glutaminolysis in cancer cells (35). It was demonstrated that endogenous c-MYC increased the turnover of the TCA cycle (36). The abnormal expression of c-MYC can drive aerobic glycolysis and oxidative phosphorylation in the meantime. Both c-MYC and HIF1 can synergistically promote the transcription of genes related to aerobic glycolysis, such as GLUT1, HK, PFKP, PKM2, LDHA, and ABC transporters, and then inhibit their protein expression, so as to promote aerobic glycolysis, establish the Warburg effect, promote the production of ATP, and accelerate the progression of tumors (34,37). Therefore, variation in cellular energy metabolism mediated by c-MYC and HIF1 may be another target for cancer therapy.
However, research on small molecule inhibitors of the c-MYC protein is facing difficulties. The c-MYC protein has no site in which small molecules can easily bind, complicating the design of direct inhibitors. The indirect targeting strategies based on the bromodomain-containing protein 4 (BRD4) which can regulate its gene transcription and the aurora kinase A (AURKA) stabilizing c-MYC need to be further studied. Some small molecule inhibiters focusing on HIF and HIF2 have been discovered, but there is still a lack of small molecule inhibitors for HIF1. The suggestion that regulating gene transcription and then further inhibiting protein expression based on the special secondary structure of nucleic acids may provide a new strategy for tumor prevention and treatment targeting c-MYC and HIF1α proteins.
The G-quadruplex structure of c-MYC and its small molecule ligands have been widely investigated. The HIF1α G-quadruplex has also been reported (27), but there are few studies on its binding with small molecule drugs. In this study, both the selected c-MYC and HIF1α G-quadruplex sequences showed parallel conformations and could combine with berberine in vitro. Interestingly, they had different binding characteristics with berberine. UV-Vis absorption and fluorescence emission spectra showed that HIF1α G-quadruplex had a stronger binding ability with berberine compared to c-MYC G-quadruplex, which was more affected by temperature. The differences in stoichiometric ratios and the thermal stability of c-MYC and HIF1α G-quadruplexes combined with berberine also indicated that the binding capacity of berberine with c-MYC G-quadruplex was different from that with HIF1α G-quadruplex. Interacting with the G-quadruplex structure may be one of the anticancer mechanisms of berberine, and the different effects of berberine on the G-quadruplexes of different genes may determine its function on each gene target.
If so, what biological effects would be induced when berberine is combined with c-MYC and HIF1α G-quadruplexes in colon cancer cells? We preliminarily proved that berberine could inhibit cell proliferation, promote cell apoptosis, and suppress gene transcription of c-MYC and HIF1α simultaneously, as well as their protein expression. Through transcriptome sequencing and bioinformatics analysis, we predicted the possible metabolic regulatory pathways of berberine, including pathways associated with c-MYC and HIF1α such as glycolysis/gluconeogenesis and TCA cycle. The results implied that berberine might exert anticancer effects partly by regulating glycolysis/gluconeogenesis and TCA cycle metabolism pathways based on stabilizing the structures of c-MYC and HIF1α G-quadruplexes at the same time.
Animal experiments were further performed to confirm the effects of berberine on colon cancer based on the targets of c-MYC and HIF1α. Berberine inhibited the protein expression of c-MYC and HIF1α while delaying the tumor growth of colon cancer mice. The targeted metabolomics study of central carbon showed that berberine reduced the concentrations of ATP, ADP and phosphoenolpyruvate, and increased the concentrations of cis-aconitate, isocitrate, homogentisate, 3-ureidopropionate and quinolinic acid in tumor tissues. ATP is an important substance in the process of energy metabolism and participates in multiple metabolic pathways such as the TCA cycle, pentose phosphate pathway, glycolysis, and gluconeogenesis. ATP can produce ADP and phosphate under the action of hydrolase. During tumorigenesis, TCA cycle is inhibited and aerobic glycolysis is promoted. However, with the presence of berberine, TCA cycle was promoted and aerobic glycolysis was inhibited. Nicotinate and nicotinamide metabolism, ubiquinone and other terpenoid-quinone biosynthesis, and pantothenate and CoA biosynthesis can provide materials for biological redox reactions. Thus it can be seen that berberine ultimately reduce ATP through regulating TCA cycle, glycolysis/gluconeogenesis and so on based on the expression inhibition of c-MYC and HIF1α in tumor tissues, which may be caused by berberine interacting with the G-quadruplex in c-MYC and HIF1α promoters, as shown in Figure 7. Furthermore, berberine could also inhibit colon cancer by regulating purine metabolism and other metabolism, but whether they depend on the targets of c-MYC and HIF1α requires further research. Thirteen common metabolic pathways were enriched by central carbon metabolism in mice and human cell transcriptome sequencing, including TCA cycle and glycolysis/gluconeogenesis, indicating the reliability of the research.
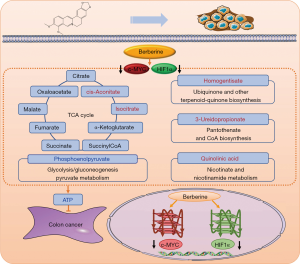
In summary, berberine could bind with c-MYC and HIF1α G-quadruplexes, but the interactions were different. The interactions both involved π-π stacking. There were 3 stoichiometric ratios for berberine combined with c-MYC, and only 1 stoichiometric ratio for berberine binding with HIF1α, suggesting that the binding mode of berberine with c-MYC was more complex, while that of berberine with HIF1α was unique. The combination of berberine and c-MYC was more susceptible to temperature compared to that of berberine and HIF1α. Furthermore, c-MYC G-quadruplex had better thermal stability than HIF1α G-quadruplex, and berberine could improve the thermal stability of both of them. On this foundation, berberine could simultaneously inhibit gene transcription of c-MYC and HIF1α as well as protein expression in colon cancer HCT116 cells. In vivo, berberine inhibited the protein expression of c-MYC and HIF1α in tumor tissues, regulated metabolic pathways such as the TCA cycle and glycolysis/gluconeogenesis, and reduced the concentration of ATP, in order to inhibit tumor progression in colon cancer mice. The G-quadruplex structures in the promoters of c-MYC and HIF1α may be potential targets for anticancer drugs regulating metabolism in colon cancer. Of course, there are some limitations: due to the large individual differences and small sample size, the concentration standard deviation of some metabolites was large; only one kind of colon cancer animal model (transplanted tumor mice) was adopted, the results were not verified in other colon cancer models such as colitis-associated colon cancer mice and Apcmin/+ mice. Therefore, further studies based on large sample size and different animal models are needed.
Acknowledgments
Funding: This work was sponsored by the National Natural Science Foundation of China (No. 81803773).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-389/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-389/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-389/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animal experiment was approved by the Scientific Research Ethics Committee of Beijing Shijitan Hospital, Capital Medical University (Project No. 2018 scientific research ethics review No. 70). The animal experiment protocol followed the guidelines of China legislations on the ethical use and care of laboratory animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Burnett-Hartman AN, Lee JK, Demb J, et al. An Update on the Epidemiology, Molecular Characterization, Diagnosis, and Screening Strategies for Early-Onset Colorectal Cancer. Gastroenterology 2021;160:1041-9. [Crossref] [PubMed]
- Akimoto N, Ugai T, Zhong R, et al. Rising incidence of early-onset colorectal cancer - a call to action. Nat Rev Clin Oncol 2021;18:230-43. [Crossref] [PubMed]
- Wei FZ, Mei SW, Wang ZJ, et al. Differential Expression Analysis Revealing CLCA1 to Be a Prognostic and Diagnostic Biomarker for Colorectal Cancer. Front Oncol 2020;10:573295. [Crossref] [PubMed]
- Zhang M, Wang HZ, Peng RY, et al. Metabolism-Associated Molecular Classification of Colorectal Cancer. Front Oncol 2020;10:602498. [Crossref] [PubMed]
- Oh HH, Joo YE. Novel biomarkers for the diagnosis and prognosis of colorectal cancer. Intest Res 2020;18:168-83. [Crossref] [PubMed]
- Hutton JE, Wang X, Zimmerman LJ, et al. Oncogenic KRAS and BRAF Drive Metabolic Reprogramming in Colorectal Cancer. Mol Cell Proteomics 2016;15:2924-38. [Crossref] [PubMed]
- Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell Mol Life Sci 2008;65:3981-99. [Crossref] [PubMed]
- Dejure FR, Eilers M. MYC and tumor metabolism: chicken and egg. EMBO J 2017;36:3409-20. [Crossref] [PubMed]
- Carvalho J, Mergny JL, Salgado GF, et al. G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies. Trends Mol Med 2020;26:848-61. [Crossref] [PubMed]
- Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet 2012;13:770-80. [Crossref] [PubMed]
- Li F, Tan W, Chen H, et al. Up- and downregulation of mature miR-1587 function by modulating its G-quadruplex structure and using small molecules. Int J Biol Macromol 2019;121:127-34. [Crossref] [PubMed]
- D'Aria F, D'Amore VM, Di Leva FS, et al. Targeting the KRAS oncogene: Synthesis, physicochemical and biological evaluation of novel G-Quadruplex DNA binders. Eur J Pharm Sci 2020;149:105337. [Crossref] [PubMed]
- Volná A, Bartas M, Karlický V, et al. G-Quadruplex in Gene Encoding Large Subunit of Plant RNA Polymerase II: A Billion-Year-Old Story. Int J Mol Sci 2021;22:7381. [Crossref] [PubMed]
- Roxo C, Kotkowiak W, Pasternak A. G4 Matters-The Influence of G-Quadruplex Structural Elements on the Antiproliferative Properties of G-Rich Oligonucleotides. Int J Mol Sci 2021;22:4941. [Crossref] [PubMed]
- Tan W, Yi L, Zhu Z, et al. Hsa-miR-1587 G-quadruplex formation and dimerization induced by NH4+, molecular crowding environment and jatrorrhizine derivatives. Talanta 2018;179:337-43. [Crossref] [PubMed]
- Huang MC, Chu IT, Wang ZF, et al. A G-Quadruplex Structure in the Promoter Region of CLIC4 Functions as a Regulatory Element for Gene Expression. Int J Mol Sci 2018;19:2678. [Crossref] [PubMed]
- Wang W, Hu S, Gu Y, et al. Human MYC G-quadruplex: From discovery to a cancer therapeutic target. Biochim Biophys Acta Rev Cancer 2020;1874:188410. [Crossref] [PubMed]
- Ferino A, Marquevielle J, Choudhary H, et al. hnRNPA1/UP1 Unfolds KRAS G-Quadruplexes and Feeds a Regulatory Axis Controlling Gene Expression. ACS Omega 2021;6:34092-106. [Crossref] [PubMed]
- Sanchez-Martin V, Lopez-Pujante C, Soriano-Rodriguez M, et al. An Updated Focus on Quadruplex Structures as Potential Therapeutic Targets in Cancer. Int J Mol Sci 2020;21:8900. [Crossref] [PubMed]
- Habtemariam S. Berberine pharmacology and the gut microbiota: A hidden therapeutic link. Pharmacol Res 2020;155:104722. [Crossref] [PubMed]
- Zhou M, Deng Y, Liu M, et al. The pharmacological activity of berberine, a review for liver protection. Eur J Pharmacol 2021;890:173655. [Crossref] [PubMed]
- Wang S, Xu Z, Cai B, et al. Berberine as a Potential Multi-Target Agent for Metabolic Diseases: A Review of Investigations for Berberine. Endocr Metab Immune Disord Drug Targets 2021;21:971-9. [Crossref] [PubMed]
- Tong M, Liu H, Hao J, et al. Comparative pharmacoproteomics reveals potential targets for berberine, a promising therapy for colorectal cancer. Biochem Biophys Res Commun 2020; Epub ahead of print. [Crossref] [PubMed]
- Kwon S, Chan AT. Extracting the benefits of berberine for colorectal cancer. Lancet Gastroenterol Hepatol 2020;5:231-3. [Crossref] [PubMed]
- Chen YX, Gao QY, Zou TH, et al. Berberine versus placebo for the prevention of recurrence of colorectal adenoma: a multicentre, double-blinded, randomised controlled study. Lancet Gastroenterol Hepatol 2020;5:267-75. [Crossref] [PubMed]
- Hallajzadeh J, Maleki Dana P, Mobini M, et al. Targeting of oncogenic signaling pathways by berberine for treatment of colorectal cancer. Med Oncol 2020;37:49. [Crossref] [PubMed]
- Lombardo CM, Welsh SJ, Strauss SJ, et al. A novel series of G-quadruplex ligands with selectivity for HIF-expressing osteosarcoma and renal cancer cell lines. Bioorg Med Chem Lett 2012;22:5984-8. [Crossref] [PubMed]
- Wen LN, Xie MX. Spectroscopic investigation of the interaction between G-quadruplex of KRAS promoter sequence and three isoquinoline alkaloids. Spectrochim Acta A Mol Biomol Spectrosc 2017;171:287-96. [Crossref] [PubMed]
- Dickerhoff J, Brundridge N, McLuckey SA, et al. Berberine Molecular Recognition of the Parallel MYC G-Quadruplex in Solution. J Med Chem 2021;64:16205-12. [Crossref] [PubMed]
- Wang Z, Jiang L, Wang J, et al. Morphine promotes angiogenesis by activating PI3K/Akt/HIF-1α pathway and upregulating VEGF in hepatocellular carcinoma. J Gastrointest Oncol 2021;12:1761-72. [Crossref] [PubMed]
- Li H, Xing X, Zhang X, et al. Effects of triptolide on the sphingosine kinase - Sphingosine-1-phosphate signaling pathway in colitis-associated colon cancer. Int Immunopharmacol 2020;88:106892. [Crossref] [PubMed]
- Zhu H, Cao C, Wu Z, et al. The probiotic L. casei Zhang slows the progression of acute and chronic kidney disease. Cell Metab 2021;33:1926-1942. [Crossref] [PubMed]
- Gupta S, Roy A, Dwarakanath BS. Metabolic Cooperation and Competition in the Tumor Microenvironment: Implications for Therapy. Front Oncol 2017;7:68. [Crossref] [PubMed]
- Marbaniang C, Kma L. Dysregulation of Glucose Metabolism by Oncogenes and Tumor Suppressors in Cancer Cells Asian Pac J Cancer Prev 2018;19:2377-90. [PubMed]
- Chen JQ, Russo J. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells. Biochim Biophys Acta 2012;1826:370-84. [PubMed]
- Morrish F, Isern N, Sadilek M, et al. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene 2009;28:2485-91. [Crossref] [PubMed]
- Liu C, Jin Y, Fan Z. The Mechanism of Warburg Effect-Induced Chemoresistance in Cancer. Front Oncol 2021;11:698023. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


