
LINC00857 promotes the proliferation of pancreatic cancer via MET, STAT3, and CREB
Introduction
Pancreatic cancer (PC) is one of the most common types of cancer. The 5-year survival rate is only 10%, and the morbidity and mortality of PC has been increasing over the years (1). Currently, targeted therapies are substantially improving the quality of life of patients with various types of cancer and prolonging their survival. However, progress in targeted therapy depends on the discovery of new targets, and not much previous work has been done in this area for PC.
Long non-coding RNAs (lncRNAs), which are over 200 nucleotides in length, are a class of RNAs with limited or no functionality in encoding proteins (2). Recent studies have highlighted the crucial role of lncRNAs in the growth, development, differentiation, proliferation, apoptosis, and metastasis of cancer cells (3-6). Moreover, studies have revealed that the dysregulation of lncRNAs is correlated with the occurrence, progress, metastasis, and prognosis of various cancers (7). LncRNAs have been shown to be significant in the pathogenesis of PC and are considered promising candidate biomarkers of PC (8). However, the functions and mechanisms of lncRNAs have yet to be fully clarified, and their exact role in PC is still under investigation (9). Currently, there are few published studies on the influence of lncRNAs on PC. Sun et al. (10) reported a substantial decrease of the lncRNA ENST00000480739 in the tumor tissues of PC, and the knockdown of this lncRNA promoted the invasion of PC. Zhao et al. (11) reported that the plasmacytoma variant translocation 1 (PVT1) is significantly overexpressed in PC tissues, and silencing PVT1 in PC cells inhibited their proliferation and migration. These discoveries further demonstrated that lncRNAs are potential biomarkers of PC, but their functions and the consequences of abnormal expression are yet to be well recorded.
In the present study, we firstly derived the LINC00857 data from GEPIA website, which predicted that LINC00857 was upregulated in PC and associated with good prognosis in PC patients. Recent studies have reported that lncRNA LINC00857 is involved in the tumorigenesis of lung cancer (12), gastric cancer (13), hepatic cancer (14), esophageal cancer (15), and ovarian cancer (16). Recently, it has been reported that LINC00857 is significantly upregulated in PC cell lines, and LINC00857 has been identified as a tumorigenic lncRNA in PC (17). However, the physiological function of LINC00857 in the progression of PC is still largely unclear, especially the downstream molecules.
In this study, we further explored the biological function and potential molecular mechanism of LINC00857 in PC. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-723).
Methods
Cell culture
The PC cell lines BxPc3 and PANC1 were purchased from Sigma-Aldrich (St Louis, MO, USA). All media were supplemented with 10% fetal bovine serum (Invitrogen, San Diego, CA, USA).
RNA isolation, reverse transcription polymerase chain reaction (RT-PCR), and quantitative PCR (qPCR)
The QIAGEN miRNeasy Mini kit (QIAGEN, Hilden, Germany) was used to extract total RNA from PC tissues and cultured cells following the manufacturers' instructions. Reverse transcription was conducted using the High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Waltham, MA, USA). The resultant complementary DNA (cDNA) was used as a template for subsequent qPCR, which was performed using Power SYBR Green Master Mix (Thermo Fisher Scientific), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an endogenous control, and the StepOne Real-Time PCR System (Thermo Fisher Scientific) to determine the relative RNA level. All data were analyzed using the 2‑ΔΔCq method.
Cell proliferation assays
Cell proliferation assays were used to evaluate the proliferation ability of PC cell lines. After small interfering RNA (siRNA) transfection for 48 hours, we placed the cells into wells of a 96-well plate (density 1×103 cells/well) and evaluated the rate of proliferation of these cells using a WST-1 assay (reagents purchased from Roche, Mannheim, Germany).
Antibodies and western blotting
Cells were harvested 72 hours after siRNA transfection as described previously (18). Western blotting was then performed according to the standard protocol, with specific anti-human antibodies mesenchymal-epithelial transition (MET), signal transducer and activator of transcription 3 (STAT3), cAMP response element-binding protein (CREB) (Cell Signaling Technology, Danvers, MA, USA), and GAPDH (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA).
Statistical analysis
SPSS standard V.16.0 and GraphPad Prism 7 were used for data analysis. The unpaired Student’s t-test was used to evaluate cell growth rate. All values are presented as mean ± standard deviation (SD). Independent triplicated experiments were performed and the data are presented as the mean ± SD. P<0.05.
Results
The expression level of LINC00857 in PC according to online database
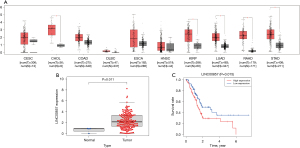
We used Gene Expression Profiling Interactive Analysis (GEPIA) to investigate transcriptional data of LINC00857 in patients with different cancers. The expression of LINC00857 was higher in many cancer tissues compared with the corresponding normal tissue (Figure 1A). According to data from GEPIA and The Cancer Genome Atlas (TCGA), the expression level of LINC00857 in PC tissue was significantly higher than that in normal tissue (P<0.05, Figure 1A and P=0.01, Figure 1B, respectively). Based on data from TCGA, the overall survival of PC patients with low expression of LINC00857 was significantly longer than that of patients with high expression (P=0.02, Figure 1C).
Cell proliferation was decreased after knockdown of LINC00857 in PC cells
To examine the level of LINC00857 expression in PC cells, RT-PCR was carried out on 5 cell lines: 4 tumor cell lines (BxPc3, PANC1, PCI-35, and Capan-1) and 1 normal cell line [human pancreatic duct epithelial (HPDE)]. The results revealed that LINC00857 was indeed overexpressed in the tumor cell lines (*, P<0.05, Figure 2A). The experiments that followed were carried out on BxPc3 and PANC1 cell lines only.
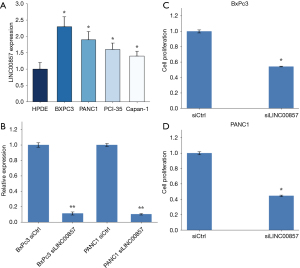
To examine the function of LINC00857, we established LINC00857 knockdown and control cell lines. We performed siRNA-mediated knockdown in BxPc3 and PANC1 cell lines, which contain high levels of the endogenous LINC00857 transcript. The expression of LINC00857 was significantly reduced after transfection with LINC00857 siRNAs, as determined by RT-PCR assays (Figure 2B). We performed the WST-1 assay to investigate the impact of LINC00857 on cell proliferation. The proliferation of BxPc3 and PANC1 cells was significantly inhibited (P<0.05) after LINC00857 knockdown (Figure 2C,2D). The results suggested that LINC00857 promoted the proliferation of PC cells.
Potential molecular signaling regulated by LINC00857
We detected protein and messenger RNA (mRNA) expression in BxPc3 and PANC1 cell lines to explore the downstream molecules of LINC00857. We applied western blot to identify proteins whose expression altered after LINC00857 knockdown and we found that in both cell lines, the protein expression of MET, STAT3, and CREB was downregulated after LINC00857 knockdown (Figure 3A). The results indicated that these downregulated proteins might have played an important role in regulating cell growth in PC cells in the LINC00857 network.

To identify whether the RNAs transcribing these proteins were regulated by LINC00857, we then detected the RNA expression of MET, STAT3, and CREB in siLINC00857 cell lines by RT-PCR (Figure 3B). We found that MET mRNA was downregulated in both BxPc3 and PANC1 cells, which was consistent with the western blot results, indicating that the MET gene was regulated at the transcriptional level by LINC00857. Meanwhile, the mRNA level of STAT3 and CREB was increased after LINC00857 knockdown, which was contrary to the western blot results, suggesting that these proteins were regulated at the posttranscriptional level.
Conversely, the protein expression of STAT3 and CREB was downregulated in PC cells with MET knockdown (Figure 3A). We conducted RT-PCR to identify whether the RNAs of LINC00857, STAT3, and CREB were regulated by MET. It was observed that the RNA expression of STAT3 and LINC00857 was upregulated, while that of CREB was downregulated after the knockdown of MET in BxPc3 cells. However, in the PANC1 cell lines that received the same treatment, the RNA expression level of LINC00857 and STAT3 was downregulated while that of CREB was upregulated (Figure 3C). In short, the transcription level changes in the 2 cell lines were exactly the opposite, implying that MET does not regulate LINC00857. Combining this with the results presented in Figure 3A, we deduced that the position of MET in the signaling pathway lies downstream of LINC00857, and that the regulating effect of MET on STAT3 and CREB are at the posttranscriptional level instead of at the transcriptional level.
In order to further clarify the relationship among LINC00857, MET, STAT3, and CREB proteins and their roles in PC proliferation, we performed MET siRNA knockdown on BxPc3 and PANC1 cells. The knockdown efficiency is shown in Figure 4A. After MET knockdown, cell proliferation measured by WST-1 assay was significantly decreased relative to the control group (P<0.05) (Figure 4B,4C). This suggested that MET was an important oncogene in LINC00857 regulating cancer progression in PC cells.

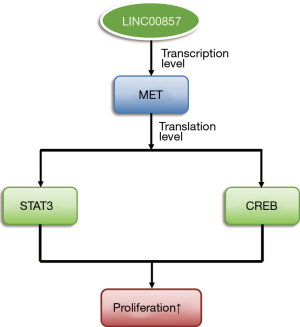
Taken together, the effect of LINC00857 on tumor cell proliferation may be via MET, STAT3, and CREB oncoproteins. A diagram of the mechanism is shown in Figure 5.
Discussion
In recent years, the relationship between lncRNAs and tumor growth has attracted substantial attention from researchers. It has been reported that lncRNAs are aberrantly expressed in various types of cancers and may be biomarkers of early diagnosis and prognosis, as well as potential targets for therapies. Researchers are currently identifying more and more types of lncRNAs that are aberrantly expressed in PC. These lncRNAs participate in tumorigenesis and play a crucial role in the development of PC (19-21).
LINC00857 is a lncRNA transcribed from the chromosome 11q22.3. Recent studies have shown that LINC00857 promoted cell proliferation and migration via the regulation of several genes related to cell survival and the cell cycle (22). LINC00857 is one of the most upregulated lncRNAs in lung cancer (23), while the silencing of LINC00857 was found to enhance radiosensitivity and promote apoptosis in lung adenocarcinoma (LUAD) cells and suppress their proliferation (22). Further, the knockdown of LINC00857 has been reported to suppress epithelial-mesenchymal transition (EMT) in hepatocellular carcinoma (HCC) (14). Our study revealed a significant upregulation of LINC00857 in PC cell lines. In previous studies, it has been reported that the overexpression of LINC00857 in PC patients was significantly correlated to their mortality (24). We found that knockdown of LINC00857 with siRNA in PC cell lines led to a decrease in cell proliferation, indicating that LINC00857 might have been involved in the growth of PC. The findings presented above suggest that LINC00857 may act as an oncogene and play an important role in the progression of PC.
In this study, we found that several oncogenic proteins, including MET, STAT3, and CREB, are downregulated after the knockdown of LINC00857. MET, as a widely known oncogene, has been confirmed to promote the proliferation of cancer cells in several types of cancers, including lung cancer (25), gastric cancer (26), and hepatic cancer (27). Studies to date have suggested that MET can activate a number of cellular signaling pathways, and therefore promote proliferation, survival, invasion, migration, angiogenesis, and EMT, and consequently, the propagation of cancer (28). Since MET is an essential oncogene in many types of tumors, therapies targeting c-MET may be an effective strategy for patients with c-MET variations (29). We revealed that after the knockdown of LINC00857, MET protein and mRNA were both decreased, suggesting that LINC00857 regulated MET expression at the transcriptional level. LINC00857 has also been found to play an oncogenic role by regulating the expression of MET in other cancers. For instance, the knockdown of LINC00857 was found to suppress the proliferation of esophageal cancer cells via the MET oncogenic protein (15), and the interaction between LINC00857 and YBX1 has been shown to regulate apoptosis and autophagy via MET (12). Moreover, similar effects have been observed in other types of lncRNAs. Su et al. discovered that the knockdown of the lncRNA MIR22HG triggered cellular survival/death via the oncogene MET (18). In this study, we determined that LINC00857 regulated the expression of MET at the transcriptional level.
STAT3 serves as both a transcription activator and an oncogene that is strictly regulated under normal physiological conditions. There is substantial evidence to support that STAT3 is sustainedly activated in most human cancer cell lines and tumor tissues, and that it is involved in processes that promote the development of cancer, such as proliferation, invasion, and migration of tumor cells. The overexpression of STAT3 is significantly correlated to a poor tumor prognosis (30-33). Previous studies have shown that the activation of STAT3 promoted the stemness and tumorigenicity in PC cells (34). In the treatment of PC, sustained activation of STAT3 signaling leads to resistance to gemcitabine, a first-line chemotherapy medication (35). Targeted treatment aimed at STAT3 can improve the sensitivity of PC to gemcitabine and enhance the efficacy of treatment (36). Currently, therapy targeting STAT3 is receiving widespread attention as a potential strategy for treating cancer and rendering cancer cells sensitive to chemotherapies. However, the signaling pathway network in which STAT3 acts as a downstream effector is very complex, and its specificity and mechanism of upstream regulation is yet to be fully clarified. Our study revealed that LINC00857 regulated the expression of STAT3 at the post-transcriptive level.
CREB is a transcription factor whose expression and functionality has been confirmed to play a significant role in cancer. CREB reinforces the expression of many target genes that are involved in various cellular functions, including survival, metabolism, and DNA repair, and is correlated to the overall survival of patients as well as treatment efficacy (37). CREB is activated via the phosphorylation of several critical oncogenic pathways, such as protein kinase A (PKA), and phosphorylated CREB (pCREB) drives the progression of cancer by recruiting additional components of the transcriptional mechanism (38). A previous study showed that elevated pCREB was strongly correlated with a worse overall survival rate in PC patients (39). Given the important effect of CREB in cancer progression, its inhibition has increasingly become an attractive strategy in the treatment of cancer; currently, there are several CREB regulators being developed as chemical probes for cancer treatment (40). In this study, we confirmed that the knockdown of LINC00857 lowered the protein expression of CREB.
A potential limitation of the present study is that LINC00857 function was not investigated in an animal model of PC. However, in vivo experiments will be performed in future investigations to further confirm the findings from the present study and to validate the accuracy and effectiveness of LINC00857 as a representative biomarker for PC.
Overall, our study revealed that the expression level of LINC00857 in PC tissue was significantly higher than that in normal tissue and the overall survival of PC patients with low expression of LINC00857 was significantly longer than that of patients with high expression. We elucidated that LINC00857 promoted cancer development in PC cancer cells. LINC00857 might have affected the proliferation of tumor cells via the oncogenic proteins MET, STAT3, and CREB, which might bring a different insight into understanding the mechanisms underlying PC pathogenesis. These findings further confirm the potential of LINC00857 as a biomarker in diagnosis and as a target in therapies.
Acknowledgments
Funding: This work was supported in part by the National Natural Science Foundation of China (NSFC) (Grant No. 81702270 to FN) and the Clinical Key Specialty Construction Project of Guangzhou Medical University (Grant No. YYPT202017).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/jgo-21-723
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-723
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-723). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer 2018;18:5-18. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet 2020;21:102-17. [Crossref] [PubMed]
- Wang H, Huang C, Yao X. The functions of long non-coding RNAs in colorectal cancer. Transl Cancer Res 2019;8:2192-204. [Crossref]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Sharma GG, Okada Y, Von Hoff D, et al. Non-coding RNA biomarkers in pancreatic ductal adenocarcinoma. Semin Cancer Biol 2021;75:153-68. [Crossref] [PubMed]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651-69. [Crossref] [PubMed]
- Sun YW, Chen YF, Li J, et al. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1α in pancreatic ductal adenocarcinoma. Br J Cancer 2014;111:2131-41. [Crossref] [PubMed]
- Zhao L, Kong H, Sun H, et al. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol 2018;233:4044-55. [Crossref] [PubMed]
- Su W, Wang L, Zhao H, et al. LINC00857 Interacting with YBX1 to Regulate Apoptosis and Autophagy via MET and Phosphor-AMPKa Signaling. Mol Ther Nucleic Acids 2020;22:1164-75. [Crossref] [PubMed]
- Pang K, Ran MJ, Zou FW, et al. Long non-coding RNA LINC00857 promotes gastric cancer cell proliferation and predicts poor patient survival. Oncol Lett 2018;16:2119-24. [Crossref] [PubMed]
- Xia C, Zhang XY, Liu W, et al. LINC00857 contributes to hepatocellular carcinoma malignancy via enhancing epithelial-mesenchymal transition. J Cell Biochem 2018; [Epub ahead of print]. [PubMed]
- Su W, Wang L, Niu F, et al. LINC00857 knockdown inhibits cell proliferation and induces apoptosis via involving STAT3 and MET oncogenic proteins in esophageal adenocarcinoma. Aging (Albany NY) 2019;11:2812-21. [Crossref] [PubMed]
- Lin X, Feng D, Li P, et al. LncRNA LINC00857 regulates the progression and glycolysis in ovarian cancer by modulating the Hippo signaling pathway. Cancer Med 2020;9:8122-32. [Crossref] [PubMed]
- Meng X, Deng Y, He S, et al. m6A-Mediated Upregulation of LINC00857 Promotes Pancreatic Cancer Tumorigenesis by Regulating the miR-150-5p/E2F3 Axis. Front Oncol 2021;11:629947. [Crossref] [PubMed]
- Su W, Feng S, Chen X, et al. Silencing of Long Noncoding RNA MIR22HG Triggers Cell Survival/Death Signaling via Oncogenes YBX1, MET, and p21 in Lung Cancer. Cancer Res 2018;78:3207-19. [Crossref] [PubMed]
- Lv Y, Huang S. Role of non-coding RNA in pancreatic cancer. Oncol Lett 2019;18:3963-73. [Crossref] [PubMed]
- Wang J, He Z, Xu J, et al. Long noncoding RNA LINC00941 promotes pancreatic cancer progression by competitively binding miR-335-5p to regulate ROCK1-mediated LIMK1/Cofilin-1 signaling. Cell Death Dis 2021;12:36. [Crossref] [PubMed]
- Zhu J, Zhu S, Yu Q, et al. LncRNA FAM66C inhibits pancreatic cancer progression by sponging miR-574-3p. Transl Cancer Res 2020;9:1806-17. [Crossref]
- Han F, Yang S, Wang W, et al. Silencing of lncRNA LINC00857 Enhances BIRC5-Dependent Radio-Sensitivity of Lung Adenocarcinoma Cells by Recruiting NF-κB1. Mol Ther Nucleic Acids 2020;22:981-93. [Crossref] [PubMed]
- Wang L, He Y, Liu W, et al. Non-coding RNA LINC00857 is predictive of poor patient survival and promotes tumor progression via cell cycle regulation in lung cancer. Oncotarget 2016;7:11487-99. [Crossref] [PubMed]
- Li T, Zhao H, Zhou H, et al. LncRNA LINC00857 strengthens the malignancy behaviors of pancreatic adenocarcinoma cells by serving as a competing endogenous RNA for miR-340-5p to upregulate TGFA expression. PLoS One 2021;16:e0247817. [Crossref] [PubMed]
- Awad MM, Oxnard GR, Jackman DM, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol 2016;34:721-30. [Crossref] [PubMed]
- Kawakami H, Okamoto I. MET-targeted therapy for gastric cancer: the importance of a biomarker-based strategy. Gastric Cancer 2016;19:687-95. [Crossref] [PubMed]
- Matsumoto K, Umitsu M, De Silva DM, et al. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci 2017;108:296-307. [Crossref] [PubMed]
- Drilon A, Cappuzzo F, Ou SI, et al. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol 2017;12:15-26. [Crossref] [PubMed]
- Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov 2008;7:504-16. [Crossref] [PubMed]
- Ying H, Da L, Yu-xiu S, et al. TLR4 mediates MAPK-STAT3 axis activation in bladder epithelial cells. Inflammation 2013;36:1064-74. [Crossref] [PubMed]
- Fu XQ, Liu B, Wang YP, et al. Activation of STAT3 is a key event in TLR4 signaling-mediated melanoma progression. Cell Death Dis 2020;11:246. [Crossref] [PubMed]
- Kamran MZ, Patil P, Gude RP. Role of STAT3 in cancer metastasis and translational advances. Biomed Res Int 2013;2013:421821. [Crossref] [PubMed]
- Wu P, Wu D, Zhao L, et al. Prognostic role of STAT3 in solid tumors: a systematic review and meta-analysis. Oncotarget 2016;7:19863-83. [Crossref] [PubMed]
- He W, Wu J, Shi J, et al. IL22RA1/STAT3 Signaling Promotes Stemness and Tumorigenicity in Pancreatic Cancer. Cancer Res 2018;78:3293-305. [Crossref] [PubMed]
- Wörmann SM, Song L, Ai J, et al. Loss of P53 Function Activates JAK2-STAT3 Signaling to Promote Pancreatic Tumor Growth, Stroma Modification, and Gemcitabine Resistance in Mice and Is Associated With Patient Survival. Gastroenterology 2016;151:180-193.e12. [Crossref] [PubMed]
- Lankadasari MB, Aparna JS, Mohammed S, et al. Targeting S1PR1/STAT3 loop abrogates desmoplasia and chemosensitizes pancreatic cancer to gemcitabine. Theranostics 2018;8:3824-40. [Crossref] [PubMed]
- Steven A, Friedrich M, Jank P, et al. What turns CREB on? And off? And why does it matter? Cell Mol Life Sci 2020;77:4049-67. [Crossref] [PubMed]
- Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal 2004;16:1211-27. [Crossref] [PubMed]
- Srinivasan S, Totiger T, Shi C, et al. Tobacco Carcinogen-Induced Production of GM-CSF Activates CREB to Promote Pancreatic Cancer. Cancer Res 2018;78:6146-58. [Crossref] [PubMed]
- Sapio L, Salzillo A, Ragone A, et al. Targeting CREB in Cancer Therapy: A Key Candidate or One of Many? An Update. Cancers (Basel) 2020;12:3166. [Crossref] [PubMed]
(English Language Editor: A. Muijlwijk)


