
REXO4 acts as a biomarker and promotes hepatocellular carcinoma progression
Introduction
Hepatocellular carcinoma (HCC) is the most common liver cancer subtype and one of the leading causes of global cancer-related death. Rates of liver cancer are rising by over 2% per year among women (1), and HCC accounts for 10.5% and 8.3% of cancer-related mortality in men and women, respectively (2). Risk factors closely associated with HCC incidence include hepatitis B virus (HBV) infection, hepatitis C virus (HCV) infection, and alcohol intake (3). Effective treatment of this cancer type is constrained by the difficulty in detecting it in its early stages, and HCC is highly likely to recur or metastasize even after treatment (4). Many studies have explored approaches to detecting different forms of liver cancer (5), with alpha-fetoprotein (AFP) serving as a common biomarker in liver cancer screening efforts despite <70% of these tumors being positive for this protein (6). Global shifts in dietary composition and lifestyle changes have led to rising rates of nonalcoholic fatty liver disease (NAFLD) (7), and the incidence of HBV/HCV infection has been considerably reduced owing to widespread efforts to vaccinate against these viruses, thus changing the overall etiological basis of liver disease in many cases (8). Consequently, there is a clear need for the identification of new biomarkers capable of reliably detecting early-stage HCC, which has spurred a large number of studies to explore the molecular mechanisms underlying this complex disease.
RNA exonuclease 4 homolog (REXO4) is a protein that has not been thoroughly studied in the oncogenic context. Nonetheless, it has previously been linked to inflammatory pathways and to the incidence of neuropathic pain (NP), and previous study showed that REXO4 may be the promising markers of M2 polarization in NP (9). Moreover, REXO4 has been identified as having a prominent role in familial solitary pituitary adenoma (FIPA) and proved to be one of the heterozygous variants in whole-exome sequencing of affected FIPA siblings (10). Our group had confirmed its important role through bioinformatics analysis in previous research (11). To further evaluate the role of REXO4 in HCC and to thereby establish its clinical relevance, we analyzed publicly available datasets and combined our cell experiments and clinical data to explore the potential of REXO4 as a diagnostic or prognostic biomarker in HCC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-819).
Methods
REXO4 expression level
Data derived from The Cancer Genome Atlas (TCGA) and GEPIA databases were assessed to explore REXO4 expression patterns in liver cancer (12). Additionally, differences in REXO4 between HCC tumors and normal tissue samples were explored by analyzing datasets available from the Gene Expression Omnibus (GEO) database.
Cell culture and patient samples
The human HepG2, Huh7, LM3, and MHCC97H HCC cell lines and the control hepatic LO2 line were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin in a humidified 5% CO2 incubator at 37 °C.
HCC tumor and paracancerous tissue samples were collected from 50 patients in the Affiliated Hospital of Nantong University between 2018 and 2020 who had not undergone prior treatment. All patients provided informed consent to participate, and the Ethics Committee of the Affiliated Hospital of Nantong University approved this study (No. 2018-L006). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). After collection, tissue samples were snap-frozen in liquid nitrogen and stored at –80 °C.
Cell transfection
The human LM3 and Huh7 cells lines were transfected with REXO4 small interfering RNA (siRNA) or negative control (NC) (the sequences were listed in Table 1). Lipofectamine 3000 (Invitrogen) was used based on manufacture’s instruction. siRNAs were obtained from GenePharma (Shanghai, China).
Table 1
| Name | Sequences |
|---|---|
| REXO4 si-1 sense | 5'-GGAAAGGACAAAUGGUGAUTT-3' |
| REXO4 si-1 antisense | 5'-AUCACCAUUUGUCCUUUCCTT-3' |
| REXO4 si-2 sense | 5'-GCUCUGCAUAAUGACCUAATT-3' |
| REXO4 si-2 antisense | 5'-UUAGGUCAUUAUGCAGAGCGT-3' |
Cell Counting Kit-8 (CCK8)
Cell growth was assessed using the CCK8 assay. Briefly, cells (1×103 cells/wall) were seeded in 96-well plates. After 24, 48, and 72 hours, 10 μL of CCK8 solution was added to each well and incubated for 1 hour before the absorbance was measured at 450 nm.
Wound-healing assay
A scratch wound was generated in confluent LM3 and Huh7 cell monolayers in a 6-well plate with a sterile 100-μL pipette tip. The wound site was imaged at 0, 24, 48, and 72 hours post-wounding, with the size of the wound area being quantified with Adobe Illustrator CC 2018 (Adobe, San Jose, CA, USA) to measure cellular migration.
Transwell assays
To assess the invasive and migratory activity of HCC cells, Transwell assay chambers (8-μm pore size; Corning, Corning, NY, USA) were used. For invasion assays, these Transwell chambers were coated using Matrigel (50 μL/well of a 1:8 Matrigel:DMEM solution; BD Biosciences, Franklin Lakes, NJ, USA). For migration and invasion assays, 5×104 and 3×104 cells in 200 μL of serum-free medium respectively, were added to the upper chamber, with 700 μL of media containing 20% being added to the lower chamber to serve as a chemoattractant. Cells were then incubated in these chambers for 24 or 48 hours, after which cells in the upper chamber were carefully removed with a cotton swab, with 4% paraformaldehyde being used to fix remaining migratory or invasive cells. These were then stained with crystal violet and counted via light microscopy in 5 random 100× fields of view. Analyses were repeated in triplicate.
Nude mouse subcutaneous tumor model
For the mouse model, 10 nude mice were divided into 4 groups evenly according to their body weight. After a large number of stably transfected cells had expanded, they were routinely digested, centrifuged, and counted. They were then resuspended in phosphate-buffered saline (PBS) in a 5×106/mL concentration. A syringe was used to inject 200 μL of the cell suspension into the subcutaneous area near the right forelimb of the nude mice. The state of the nude mice was recorded, and the mice were weighed. When a small induration was palpable under the skin, the length and width of the tumor were measured with a vernier caliper every 3 days, and this measurement was used to calculate the volume of the tumors. After 30 days of rearing, the mice were killed by cervical dislocation. The tumors were dissected, photographed, and weighed. All animal experiments were approved by the Committee on the Use of Live Animals in Teaching and Research, Nantong University (No. S20201125-403), in compliance with the inspection contents of national or institutional guidelines for the care and use of animals, namely the Standard Operating Procedures for Laboratory Animal Center of Nantong University. A protocol was prepared before the study without registration.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from Huh7, MHCC97H, LM3, HepG2, and LO2 cells with TRIzol (Invitrogen Life Technologies, Thermo Fisher Scientific). A RevertAid First Strand cDNA Synthesis Kit (TaKaRa Bio, Kusatsu, Japan) was used to prepare complement DNA (cDNA). All quantitative real-time polymerase chain reaction qRT-PCR assays were performed with a LightCycler 480 II instrument (Roche, Basel, Switzerland) and SYBR Master Mix (TaKaRa Bio) based on standard protocols using the following primers: REXO4: 5'-TCTCTTCCGGAGTCTTTTCCTG-3' (forward) and 5'-CCTTCACTTGAGGCGAGGTC-3' (reverse); GAPDH: (a housekeeping gene) 5'-GAAGGTGAAGGTCGGAGTC-3' (forward) and 5'-GAAGATGGTGATGGGATTTC-3' (reverse). GAPDH was used for normalization.
Western blotting analysis
Lysis buffer containing phenylmethylsulfonyl fluoride (PMSF) and radioimmunoprecipitation assay (RIPA; 10 μL PMSF/1 mL RIPA) was used to separate total protein. Next, 30 µg of protein sample was incubated with denaturation buffer (0.3 M Tris pH 6.8, 10% 2-mercaptoethanol, 40% glycerol, 20% SDS, and 0.02% bromophenol blue) at 95 °C for 5 minutes and loaded onto a 10% SDS-polyacrylamide gel for electrophoresis. Electrophoresis was first conducted at 80 V until the sample entered the separation gel, after which 120 V was used until the bromophenol blue reached the bottom of the gel. The tailored polyvinylidene fluoride (PVDF) membrane was excited in methanol for 15 s, and the target glue was cut according to the protein marker. The membrane was then transferred under the condition of a constant current of 200 mA for 30 to 120 minutes. After the transfer of the membrane, the PVDF membrane was washed with 1× tris-buffered saline with Tween20 (TBST), placed in 5% nonfat milk, and sealed in a 37 °C constant temperature shaker for 1 hour. After blocking, the PVDF membrane was washed with 1× TBST, and membranes were then incubated overnight at 4 °C in a primary antibody against REXO4 (1:1,000, 18890-1-AP, Proteintech) and GAPDH (1:2,000, sc-20357, Santa Cruz Biotechnology). The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies for 1 hour at room temperature. Immunoreactivity was detected using chemiluminescence.
TCGA RNA-sequencing data and survival
TCGA project of Genomic Data Commons (GDC) analyzes multiple human cancer samples (13). The RNA-sequencing (RNA-seq) data from 17 cancer types representing 21 cancer subtypes corresponding to the main cancer types in the human pathology map were used to compare the protein staining data from the human protein map with the RNA-seq data from TCGA database. TCGA RNA-seq data were mapped using the Ensembl gene ID provided by TCGA (14), and the fragments per kilobase of exon/million reads (FPKM) value of each gene was then used to quantify gene expression level (15). The prognosis of every group of patients was checked by Kaplan-Meier survival analysis, and the survival results of the two groups were compared by log-rank test.
Gene set enrichment analysis (GSEA)
GSEA analyses are used to determine whether there are consistent, significant differences in the expression of groups of genes between two biological states (16). For GSEA analysis, GSEA tools (http://www.broadinstitute.org/gsea) were used in this study. Gene enrichment analysis refers to the use of functional annotations or gene sets from previous experiments to sort genes according to the degree of differential expression in the two types of samples, and then to check whether the preset gene set is enriched at the top or bottom of the sorting table set (17). GSEA was used to clarify the possible REXO4-related pathways that may influence the progression of HCC.
Immune cell infiltration analyses
The gene expression data with standard annotation were uploaded to the Cell type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) web portal (http://cibersort.stanford.edu/) (18). CIBERSORT output of P<0.05 shows that the supposed scores of the immune cell population conducted by CIBERSORT are credible results, so they were considered to meet the conditions for further analysis. The final CIBERSORT output estimate is normalized to the sum of 1 for every sample, and then directly interpreted as cell fraction for comparison between different immune cell types and datasets (19).
Statistical analysis
All data are expressed as mean ± standard deviation (SD) from at least three independent experiments. Statistical analysis of differences between groups was performed with SPSS (IBM Corp., Armonk, NY, USA) or GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, USA) using Student’s paired or unpaired t-test or one-way analysis of variance. Kaplan-Meier log-rank test was used to calculate the overall survival (OS) rate. Cox proportional hazards model was used for univariate and multivariate analysis. A P value <0.05 was considered statistically significant.
Results
HCC tissue samples exhibited REXO4 upregulation
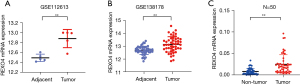
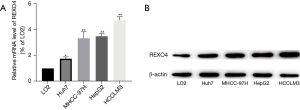
We began by assessing REXO4 messenger RNA (mRNA) expression levels in HCC tumor tissues in publicly available datasets, which revealed that this gene is upregulated in HCC tumors relative to adjacent normal tissues in the GEO datasets. Data extracted from GSE112612 and GSE138178 both showed an upregulated expression in HCC (P<0.01; Figure 1A,1B). To confirm this, we additionally analyzed the expression of REXO4 in 50 pairs of HCC tumors and paracancerous tissues from patients who underwent surgery at the Affiliated Hospital of Nantong University and obtained results consistent with those above (P<0.01, Figure 1C). For further research, we assessed REXO4 mRNA expression levels in the Huh7, MHCC-97H, Hep-G2, and HCCLM3 HCC cell lines and in the control LO2 cell line (Figure 2A). In western blotting experiments, REXO4 protein levels were found to be significantly elevated in the highly invasive LM3 and Huh7 cell lines as compared to the control LO2 cells (Figure 2B).
REXO4 upregulation was associated with a poorer HCC patient prognosis
We next explored the relationship between REXO4 expression and HCC patient clinicopathological characteristics (Table 2). Using the Gene Expression Profiling Interactive Analysis (GEPIA) database, we found that REXO4 mRNA expression was significantly increased in HCC when compared to the control group (P value <0.05, Figure 3A), suggesting that it may be a valuable diagnostic indicator for with HCC. By analyzing TCGA RNA-seq data and TCGA survival, we found that a high expression of REXO4 showed a higher number of observed events than expected events, and thus could be considered an unfavorable prognostic gene (Figure 3B). Together, these results suggested that REXO4 upregulation may be a diagnostic biomarker linked to worse HCC patient prognosis.
Table 2
| Parameters | HCC (n=45) | Low groups (n=22) | High groups (n=23) | P value |
|---|---|---|---|---|
| Age (years) | 0.763 | |||
| ≤60 | 17 | 9 | 8 | |
| >60 | 28 | 13 | 15 | |
| Gender | 0.491 | |||
| Male | 35 | 16 | 19 | |
| Female | 10 | 6 | 4 | |
| Tumor size (cm) | 0.007** | |||
| ≤5 | 25 | 17 | 8 | |
| >5 | 20 | 5 | 15 | |
| Tumor number | 0.365 | |||
| ≤1 | 27 | 15 | 12 | |
| >1 | 18 | 7 | 11 | |
| AFP (μg/L) | 0.003** | |||
| ≤20 | 20 | 15 | 5 | |
| >20 | 25 | 7 | 18 | |
| Stage | 0.047* | |||
| I–II | 37 | 21 | 16 | |
| III–IV | 8 | 1 | 7 | |
| Microvascular infiltration | 0.003** | |||
| No | 31 | 20 | 11 | |
| Yes | 14 | 2 | 12 | |
| Liver cirrhosis | 0.722 | |||
| No | 10 | 4 | 6 | |
| Yes | 35 | 18 | 17 |
*, P<0.05; **, P<0.01. HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein.

Analysis of REXO4-related signaling pathways in HCC cells
Next, we employed a GSEA approach to clarify the REXO4-regulated biological pathways involved in HCC cells. In this study, a GSEA approach was used to evaluate TCGA gene expression data from 374 liver cancer samples from UCSC Xena in order to identify biological processes associated with REXO4 (20). The number of random sample permutations was set at 1,000, and the significance threshold was set at P<0.05 (Figure 4A). These analyses revealed this gene to be closely related to fatty acid metabolism, linoleic acid metabolism, and PPAR signaling pathways, suggesting a link between REXO4 and the maintenance of lipid metabolism homeostasis in the liver (Figure 4B). We additionally analyzed the GSE138052 dataset to further expand these analyses. For this, according to the analysis of GSE138052, embryonic stem cell (ES)-derived hepatocyte-like cells were treated with lactate (10 mM), pyruvate (1 mM), and octanoate (2 mM) (LPO) to promote the accumulation of lipids within cells to emulate a phenotype consistent with NAFLD. We suspected that REXO4 may related to NAFLD related HCC (Figure 4C). We therefore conducted oil red O staining in an effort to confirm this hypothesis, which revealed increased hepatocyte lipid accumulation in regions with higher REXO4 expression (Figure 5). These findings firmly support a positive correlation between REXO4 and NAFLD-related HCC.


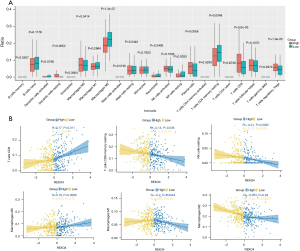
REXO4 was associated with HCC tumor immune cell infiltration
Even among patients with the same histological subtype of cancer, the characteristics of immune cell infiltration profiles may be significantly different, which may lead to distinct clinical outcomes (21). Increases in tumor-infiltrating lymphocyte numbers are generally associated with a better prognosis in HCC and many other cancer types. A considerable amount of evidence has indicated an association between the immune microenvironment and clinical outcome in HCC (22). To establish a potential link between REXO4 immune cell infiltration in HCC, we employed the CIBERSORT algorithms to assess immune cell enrichment within HCC tumor tissue samples (23). After unsupervised clustering, sample results were arranged into heat maps, and correlations analyses between immune cell infiltration and differential immune checkpoint gene expression patterns were conducted to quantify the activity or enrichment level of immune cells in HCC tumor tissues with low and high levels of REXO4 expression in the TCGA Liver Hepatocellular Carcinoma (LIHC) cohort (Figure 6A). These analyses revealed that higher levels of REXO4 expression were associated with significant increases in plasma cell, CD8+ T cell, and regulatory T cell infiltration, together with reductions in M2 macrophage, CD4+ T cell, and resting memory CD4+ T cell infiltration (Figure 6B), highlighting the differences in immune cell expression in certain LIHC cohorts. These results thus demonstrate a link between REXO4 and immune cell infiltration, indicating that this gene may shape the microenvironment within HCC tumors.
REXO4 knockdown suppressed HCC cell proliferation, invasion, and migration
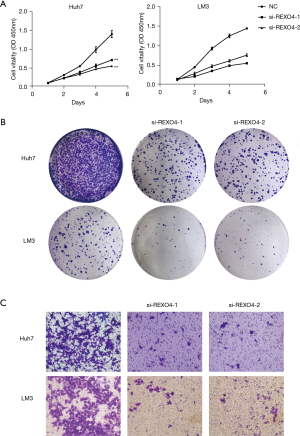
The above results together demonstrate that REXO4 might act as an oncogenic gene in HCC. We thus selected LM3 and Huh7 cells for further analyses. Given the high levels of REXO4 expression, we chose to use small interfering RNAs (siRNAs) to knockdown REXO4. When LM3 and Huh7 cells were transfected with REXO4-specific siRNAs (si-REXO4-1 and si-REXO4-2), we observed 80% and 70% reductions in REXO4 protein levels, respectively, following siREXO4-1 transfection relative to negative control (NC) (Figure 7A). Western blotting also showed the decreased expression, proving the efficiency of siRNAs (Figure 7B). These siRNAs were thus used in subsequent analyses.

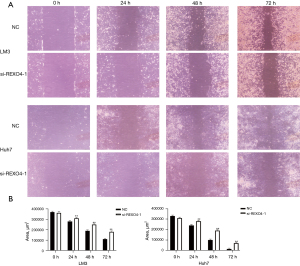
The function of REXO4 was examined with a CCK-8 assay, which confirmed that knocking down this gene significantly impaired the survival of HCC cells as compared to si-NC transfection (P<0.01; Figure 8A). A significant decrease in colony size and number in the colony formation assay relative to controls (P<0.01; Figure 8B) was observed. In Transwell assays, REXO4 silencing was additionally associated with impaired migratory and invasive activity (P<0.01; Figure 8C). Wound-healing assays measured at 0, 24, 48, and 72 hours similarly confirmed REXO4 knockdown to be associated with the suppression of primary wound closure as compared to siNC transfection, which was consistent with disrupted migration (P<0.05; Figure 9A,9B). Overall, these findings indicated that REXO4 can influence the proliferation, migration, and invasion of HCC cells.
REXO4 promoted tumor formation of liver cancer cells in nude mice
To better understand the link between REXO4 and HCC cell tumorigenesis, LM3 cells transfected with either si-REXO4-1 or a negative control construct were subcutaneously implanted into 5 male nude mice. Animals were killed 40 days later, at which time the tumor volume and weight values in the LM3-NC group were found to be significantly higher than those in the si-REXO4-1 group (P<0.01; Figure 10). These data thus further support the role of REXO4 as a promoter of HCC proliferation.

Discussion
HCC remains the most common form of liver cancer and the fourth leading cause of cancer-related death worldwide. It also incurs a massive global health burden due to its high mortality and poor prognosis, with NAFLD-related HCC becoming a topic of growing clinical concern (24). Owing to high rates of metastasis and recurrence, HCC patients experience relatively poor 5-year survival rates (25). The dearth of reliable biomarkers suitable for the early-stage-detection of HCC results in unsatisfactory curative treatment outcomes for most patients. Herein, metabolite biomarkers were identified and validated as potential correlates for HCC diagnosis. A total of 1448 subjects, including healthy controls and patients with chronic HBV infection, liver cirrhosis, and HCC, were recruited from multiple centers in China. A serum metabolite biomarker panel including phenylalanyl-tryptophan and glycocholate was defined. This validated serum metabolite biomarker panel has exhibited good diagnostic performance for the early detection of HCC in at-risk populations (13). In the past decade, there have been improvements in nondrug therapies and drug therapies for HCC treatment. Nondrug therapies include hepatic resection, liver transplantation, transarterial chemoembolization (TACE), and ablation (26). However, there are relatively few targeted treatments for HCC, and those that exist are of limited efficacy owing to intra- and intertumor heterogeneity and to the emergence of drug resistance (27). Furthermore, the mechanistic basis for HCC onset and progression is poorly understood, further complicating efforts to reliably treat this cancer type (28). The lack of specific biomarkers associated with this cancer type also represents a major obstacle to a more comprehensive understanding of this disease that must be addressed to guide effective and individualized patient treatment. While several prognostic biomarkers associated with HCC have been reported to date, a majority are not liver-specific and are of relatively limited therapeutic significance (29).
Herein, we systematically assessed the prognostic relevance of REXO4 in HCC, revealing the upregulation of this gene to be strongly correlated with poor patient outcomes. Relative to normal liver tissue samples, those from HCC patients showed significant REXO4 upregulation at the mRNA and protein levels. These REXO4 levels were additionally correlated with tumor grade, size, stage, and degree of vascular invasion as well as with the OS and disease-free survival of HCC patients. The knockdown of REXO4 in HCC cells was sufficient to impair their survival, proliferation, migration, and invasiveness to a certain extent, and REXO4 may function as an oncogenic factor.
Immune evasion, as one of the basic features of cancer, has aroused heated discussion (30). Immunotherapies, especially blockades of immune checkpoint, have showed striking performance in the treatment of malignant tumors. More and more evidence suggests immune-related components, such as immune genes and immune cells, contribute greatly to the occurrence and progression of cancer and are valuable markers for the process of cancer diagnosis and prognosis (31). In our study, GSEA further revealed this gene to be associated with lipid metabolism and the PPAR signaling pathway. We further found that REXO4 levels were highly correlated with the degree of immune cell infiltration in HCC tumors, particularly with natural killer (NK) cell and macrophage levels, suggesting a link between this gene and the susceptibility of these tumors to immunotherapeutic treatment. Our results indicated that REXO4 plays a key role in regulating immune cell recruitment and HCC progression, underscoring the need for further large-scale genomics analyses and functional studies aimed at elucidating the clinical relevance of this target in patient populations. Follow-up research should investigate the value of REXO4 as both a prognostic biomarker and therapeutic target in HCC patients.
Conclusions
In this study, we provided multiple lines of evidence supporting the key role played by REXO4 in the HCC onset and progression, highlighting its potential as a biomarker associated with this cancer type. Specifically, we found REXO4 to be related to a variety of key cancer-related activities, including lipid metabolism, that may be linked to NAFLD-related HCC, as confirmed through oil red O staining of tissue samples. In addition, REXO4 was found to be closely linked to the infiltration of NK cells and macrophages into HCC tumors, suggesting it may be an important immunoregulatory protein. Therefore, REXO4 may represent a viable target for immunotherapeutic intervention aimed at treating HCC.
Acknowledgments
We would like to thank LEXIS (Scientific Editing Experts, United States. LEXIS Academic Service, LLC) for help in polishing our manuscript.
Funding: This study was supported by grants from the National Natural Science Foundation of China (No. 81572397).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-819
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-819
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-819). All authors report they received funding from the National Natural Science Foundation of China (No. 81572397). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from the patients, and this research was approved by the Ethics Committee of Affiliated Hospital of Nantong University (No. 2018-L006). All animal experiments were approved by the Committee on the Use of Live Animals in Teaching and Research, Nantong University (No. S20201125-403), in compliance with the inspection contents of national or institutional guidelines for the care and use of animals, namely the Standard Operating Procedures for Laboratory Animal Center of Nantong University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res 2021;149:1-61. [Crossref] [PubMed]
- Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019;156:477-491.e1. [Crossref] [PubMed]
- Luo P, Yin P, Hua R, et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology 2018;67:662-75. [Crossref] [PubMed]
- Wang T, Zhang KH. New Blood Biomarkers for the Diagnosis of AFP-Negative Hepatocellular Carcinoma. Front Oncol 2020;10:1316. [Crossref] [PubMed]
- Wu Z, Ma H, Wang L, et al. Tumor suppressor ZHX2 inhibits NAFLD-HCC progression via blocking LPL-mediated lipid uptake. Cell Death Differ 2020;27:1693-708. [Crossref] [PubMed]
- Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol 2020;72:250-61. [Crossref] [PubMed]
- Krishnamurthy N, Ngam CR, Berdis AJ, et al. The exonuclease activity of hPMC2 is required for transcriptional regulation of the QR gene and repair of estrogen-induced abasic sites. Oncogene 2011;30:4731-9. [Crossref] [PubMed]
- Cai G, Zhu Y, Zhao Y, et al. Network Analysis of miRNA and mRNA Changes in the Prelimbic Cortex of Rats With Chronic Neuropathic Pain: Pointing to Inflammation. Front Genet 2020;11:612. [Crossref] [PubMed]
- Chen W, Gao C, Shen J, et al. The expression and prognostic value of REXO4 in hepatocellular carcinoma. J Gastrointest Oncol 2021;12:1704-17. [Crossref] [PubMed]
- Wang Z, Jensen MA, Zenklusen JC. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol 2016;1418:111-41. [Crossref] [PubMed]
- Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-Analyzed Tumors. Cell 2018;173:530. [Crossref] [PubMed]
- Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [Crossref] [PubMed]
- Zhang S, Liu Z, Wu D, et al. Single-Cell RNA-Seq Analysis Reveals Microenvironmental Infiltration of Plasma Cells and Hepatocytic Prognostic Markers in HCC With Cirrhosis. Front Oncol 2020;10:596318. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Ye L, Zhang T, Kang Z, et al. Tumor-Infiltrating Immune Cells Act as a Marker for Prognosis in Colorectal Cancer. Front Immunol 2019;10:2368. [Crossref] [PubMed]
- Zhou R, Zhang J, Zeng D, et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer. Cancer Immunol Immunother 2019;68:433-42. [Crossref] [PubMed]
- Li W, Chen QF, Huang T, et al. Identification and Validation of a Prognostic lncRNA Signature for Hepatocellular Carcinoma. Front Oncol 2020;10:780. [Crossref] [PubMed]
- Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018;68:1025-41. [Crossref] [PubMed]
- Sia D, Jiao Y, Martinez-Quetglas I, et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology 2017;153:812-26. [Crossref] [PubMed]
- Chen R, Zhang Y. EPDR1 correlates with immune cell infiltration in hepatocellular carcinoma and can be used as a prognostic biomarker. J Cell Mol Med 2020;24:12107-18. [Crossref] [PubMed]
- Kovalic AJ, Cholankeril G, Satapathy SK. Nonalcoholic fatty liver disease and alcoholic liver disease: metabolic diseases with systemic manifestations. Transl Gastroenterol Hepatol 2019;4:65. [Crossref] [PubMed]
- Huang DQ, El-Serag HB, Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2021;18:223-38. [Crossref] [PubMed]
- Mahara G, Chen G, Ge Q, et al. A comprehensive intervention on feasibility, efficacy, and safety between TAE combined with multi-applicator ablation therapy and TACE in the treatment of large hepatocellular carcinoma. Transl Cancer Res 2020;9:6303-12. [Crossref]
- Chang Y, Jeong SW, Young Jang J, et al. Recent Updates of Transarterial Chemoembolilzation in Hepatocellular Carcinoma. Int J Mol Sci 2020;21:8165. [Crossref] [PubMed]
- Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol 2019;12:133. [Crossref] [PubMed]
- Lee YR, Kim G, Tak WY, et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer 2019;144:1444-52. [Crossref] [PubMed]
- Tseng HC, Xiong W, Badeti S, et al. Efficacy of anti-CD147 chimeric antigen receptors targeting hepatocellular carcinoma. Nat Commun 2020;11:4810. [Crossref] [PubMed]
- Pinter M, Jain RK, Duda DG. The Current Landscape of Immune Checkpoint Blockade in Hepatocellular Carcinoma: A Review. JAMA Oncol 2021;7:113-23. [Crossref] [PubMed]
(English Language Editor: J. Gray)


