
Correlation of c-MET expression with clinical characteristics and the prognosis of colorectal cancer
Introduction
The proto-oncogene c-MET (mesenchymal-epithelial transition factor gene) encodes the tyrosine kinase receptor for hepatocyte growth factor (HGF). The interaction between HGF and c-MET plays a critical role in cellular proliferation, survival, migration, and invasion in cancers (1,2). Overexpression of c-MET is correlated with increased tumor cell motility, invasiveness, and angiogenesis, which may stimulate tumor metastatic ability and contribute to tumor aggressiveness (3). MET is considered an essential factor for early invasion and metastasis of CRC and can be regarded as an important prognostic indicator (4,5). Also, cabozantinib, a Novel c-Met Inhibitor effectively reduced tumor size and angiogenesis, and suppressed the expression of vascular endothelial growth factor (VEGF) in tumor tissues via modulating Sonic Hedgehog pathway (6). There is limited data regarding the relationship between the status of c-MET and the clinicopathological characteristics of patients with colorectal cancer (CRC), and it remains unclear whether c-MET activation is related to the status of KRAS, NRAS, and BRAF genes. Furthermore, recent studies have also suggested that activation of c-MET may be a novel mechanism of cetuximab resistance in CRC (7-9). In metastatic CRC (mCRC) patients with wild-type KRAS, MET overexpression showed a lower disease control rate (DCR) and shorter progression-free survival (PFS) compared to patients with normal MET expression (10). Immunohistochemical analysis of cMet expression in human CRC tissues showed a significant increased expression compared with matched normal tissues (11). However, it still unclear whether the expression of c-MET has any influence on survival time.
In the context of c-MET of CRC, the utility in clinical practice is not clear because its correlation with definitive specific biomarker is blurred and indistinct. So, it is urgent demand to explorer this field. So was to the attemptions of drug-resistant mCRC with the c-MET inhibitors alone, or combined. With respect to the above, we conducted a clinical study to analyze the relationship between c-MET expression and other pathological factors, and to further investigate the relationship between the level of c-MET and KRAS, NRAS, and BRAF gene status in 337 CRC patients, so as to provide new insights into clinical treatment options of anti-EGFR therapy for patients with CRC.
We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-536).
Methods
Patient population
From May 2014 to June 2015, there were 337 patients with CRC who underwent surgical resection and subsequent therapy in Guangdong General Hospital, Guangdong Academy of Medical Sciences, including mutational analysis and immunohistochemistry staining as part of their standard care. The clinical features of the patients and pathological profiles of the tumors were obtained from patient medical records. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Guangdong General Hospital (No. KY2020-420-01) and informed consent was taken from all the patients.
Immunohistochemical staining and evaluation
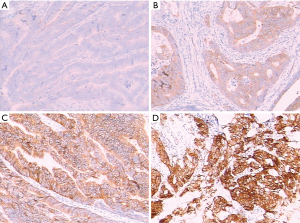
Fresh tissue samples of colorectal tumors during the operation were fixed and dehydrated. Histological paraffin sections were then prepared following paraffin embedding and sectioning for hematoxylin-eosin staining (HE staining). Microdissection was utilized under the guidance of a clinical pathologist as required to ensure >30% tumor cellularity. Immunohistochemistry of CRC samples was performed using a primary antibody against the c-MET protein with the avidin biotin procedure following the kit manufacturer’s instructions. The antibody used was a monoclonal mouse anti-human c-MET antibody (DakoCytomation Norden A/S, Glostrup, Denmark, dilution 1:100). A known positive control section for each antibody was stained under equivalent conditions to ensure proper staining. Phosphate-buffered saline (PBS) was used as a substitute for the primary antibody as a negative control in the same manner. The results for c-MET were classified into 4 scores depending on the intensity of immunostaining: 0, negative; 1, weak; 2, medium; and 3, strong (12). Scores 0 and 1 were defined as c-MET-low, and scores 2 and 3 were c-MET-high.
Mutational analysis
Tumor genotyping of all 337 patients was performed in the medical examination center of our hospital using nucleic acids extracted from diagnostic formalin-fixed paraffin-embedded tumor tissueswith the QIAGEN system (QIAGEN, Dneasy Blood and Tissue Kit, Germany). Mutational profiling simultaneously queried Kirsten rat sarcoma viral oncogene homolog (KRAS), neuroblastoma RAS viral oncogene homolog (NRAS), and v-raf murine sarcoma viral oncogene homolog B (BRAF). This was performed using a custom-modified ABI 3500DX Sequencing System (Applied Biosystems/Life Technologies Corporation, Carlsbad, American). First-generation sequencing was performed in both directions. Subsequently, using Chromas software, sequence analysis was compared with a normal sequence that was downloaded from Gene Bank.
Statistical analysis
Statistical analysis in the present study was performed using SPSS16.0 software (SPSS, Chicago, IL, USA). By applying the Chi-square test, the relationships between the immunohistochemical staining of c-MET and clinicopathological features in CRC patients who underwent surgical excision were compared (such as age at diagnosis, sex, tumor differentiation, TNM stage, metastasis, primary tumor site, lymph node metastasis). Kaplan-Meier survival curves were used to evaluate the relationships between clinicopathological features and survival outcomes in different patients with CRC. Multivariate survival analysis was further conducted with the Cox regression model to evaluate the effect of various factors on the prognosis of CRC. P<0.05 was defined as statistically significant.
Results
General information
Of the 337 patients enrolled in the present study, 192 (57.0%) were male and 145 (43.0%) were female. The median age was 62.4 years old (ranging from 23–92 years old). In terms of location, 142 cancers were from the proximal colon (cecum to transverse colon), 66 from the distal colon (descending colon to sigmoid colon), and 129 from the rectum. Meanwhile, according to the criteria of TNM staging after surgery, there were 48 patients with stage I disease, 115 patients with stage II, 111 patients with stage III, and 63 patients with stage IV. Patients were grouped into 2 groups, with 39 cases in the poor differentiation group and 298 cases in the moderate-high differentiation group. According to the metastasis status, there were 38 cases with hepatic metastasis, 20 cases with lung metastasis, and 36 cases with other multiple metastasis. Due to a high proportion of CRC patients with a high level of EGFR expression, which has been reported to have an influence on the outcomes of anti-EGFR therapy, we evaluated the patients into two groups, EGFR low expression (EGFR 0-+) and EGFR high expression (EGFR ++-+++). c-MET protein expression was revealed to have a strong correlation with the expression of EGFR (224 cases of EGFR 0-+ vs. 103 cases of EGFR ++-+++, 10 missed cases, P=0.002; Table 1). There was a significant correlation between c-MET expression and EGFR level. The details are shown in Table 1.
Table 1
| Clinical parameters | n | c-MET | P value | |
|---|---|---|---|---|
| Low-type | High-type | |||
| Gender | ||||
| Male | 192 | 141 | 51 | |
| Female | 145 | 89 | 56 | 0.016 |
| Age | ||||
| ≥65 years | 147 | 99 | 48 | |
| <65 years | 190 | 131 | 59 | 0.755 |
| Tumor location | ||||
| Right colon | 142 | 100 | 42 | |
| Left colon | 66 | 42 | 24 | 0.620 |
| Rectum | 129 | 88 | 41 | |
| Tumor size | ||||
| <5 cm | 188 | 125 | 63 | |
| ≥5 cm | 149 | 105 | 44 | 0.436 |
| Presence of nodal metastasis | ||||
| 0 Negative | 181 | 118 | 63 | |
| 1 Positive | 156 | 112 | 44 | 0.195 |
| Lymphovascular invasion | ||||
| 0 negative | 273 | 189 | 84 | |
| 1 positive | 64 | 41 | 23 | 0.425 |
| Perineural invasion | ||||
| 0 negative | 233 | 168 | 65 | |
| 1 positive | 102 | 60 | 42 | 0.017 |
| Margin status | ||||
| 0 free | 331 | 224 | 107 | |
| 1 involved | 6 | 6 | 0 | 0.092 |
| Differentiation degree | ||||
| Poor | 39 | 32 | 7 | |
| Moderate-high | 298 | 198 | 100 | 0.128 |
| Tumor stage | ||||
| I | 48 | 28 | 20 | |
| II | 115 | 80 | 35 | |
| III | 111 | 77 | 34 | |
| IV | 63 | 45 | 18 | 0.454 |
| Metastasis type | ||||
| Hepatic metastasis | 38 | 24 | 14 | 0.475 |
| Pulmonary metastasis | 20 | 13 | 7 | 0.748 |
| Other metastasis | 36 | 25 | 11 | 0.871 |
| EGFR expression | ||||
| 0 | 31 | 26 | 5 | |
| 1 | 98 | 73 | 25 | |
| 2 | 100 | 72 | 28 | |
| 3 | 98 | 53 | 45 | 0.002 |
| Missed | 10 | |||
Relationship between c-MET level and clinicopathological features in patients with CRC
c-MET expression was assessed by immunohistochemistry (IHC) in primary tumors in all 337 patients. c-MET staining intensity in the primary tumors was grade 0-1 in 230 cases (68.2%) and grade 2–3 in 107 cases (31.8%) (Figure 1). The statistical results showed that there were significant differences between gender and perineural invasion (P=0.016 and 0.017, respectively; Table 1), while there were no significant differences regarding age, tumor location, tumor size, lymphovascular invasion, margin status, metastasis, and tumor stage in different patients grouped according to different c-MET levels (P>0.05; Table 1). Of the metastatic patients included in this study, 20 cases (3.4%) had lung metastasis as the only site of metastasis, 38 cases (11.3%) had liver metastasis, 36 cases (10.7%) had other sites of metastasis, while 245 cases without metastasis were identified by c-MET level. Regarding the c-MET status, there was no significant difference in metastatic location between the 4groups (P>0.05; Table 1).
Genotyping of KRAS, NRAS, and BRAF
All 337 CRC cases were examined for mutations in KRAS (exons 2, 3, and 4), NRAS (exons 2, 3, and 3), and BRAF (exon 15). KRAS mutation was detected in 43.2% (n=146) of cases, NRAS mutation was detected in 3.3% (n=11) of cases, while BRAF mutation was detected in 4.7% (n=16) of cases. We also observed the changes in the nucleotides and corresponding amino acids of mutation profiles. The most frequent mutation of KRAS was 2p. G12D, occurring in 37 cases with a frequency of 11.0%, followed by 2p G12DV, occurring in 35 cases with a frequency of 10.4%. The most frequent NRAS mutation was 3p.61 (codon 3), occurring in 5 cases with a frequency of 1.48%, while the most frequent BRAF mutation was 15p.V600E (codon 15), occurring in 16 cases with a frequency 4.7%. The distribution of mutations was consistent with previous studies.
No association between the expression of c-MET and gene mutations of primary tumors
KRAS mutational status was verified in 146 cases, and of these, 52 cases were c-MET-high tumors while 94 cases were c-MET-low tumors, which demonstrated that c-MET protein levels of primary tumors were not associated with KRAS mutation (P=0.183). NRAS mutational status was demonstrated in 11 cases, including 2 cases with c-MET-high tumors and 9 cases with c-MET-low tumors. These data showed that c-MET protein expression was not related to NRAS mutation (P=0.326). BRAF mutational status was shown in 16 cases (6 cases of c-MET-high and 10 cases of c-MET-low), which demonstrated that c-MET expression in primary tumors had no relationship with BRAF mutation (P=0.613).
Analysis of prognostic factors in CRC patients
Patients were divided into 2 groups based on the expression level of c-MET, and they were compared in terms of baseline data. In the median follow up time of 41.5 months (from 31 to 63 months), the results of the 337 patients revealed that to the end of the study, 51 patients died and 40 patients were lost to follow up, with a median overall survival (OS) period of 28.9 months.
The relationship between c-MET expression levels and survival was also investigated. The median OS among patients with c-MET-high primary tumors was 29.2 months, while the median OS among those with c-MET-low primary tumors was 28.7 months, and the difference was not significant (P=0.379; Figure 2). The Cox proportional hazard model for multivariate survival analysis (as shown in Table 2) indicated that advanced age, NRAS mutation, advanced TNM stage, and lung metastasis were independent risk factors influencing the prognosis of patients (P=0.002, P=0.011, P=0.049, and P=0.016, respectively). Compared with younger patients (<60 years), NRAS wild-type, early TNM stage, and no lung metastasis, older patients (≥60 years), NRAS mutation, advanced TNM stage, and lung metastasis with CRC had relatively poor prognostic outcomes (Table 2).
Table 2
| Variable | B | SE | Wald | df | Sig. | Exp(B) | 95.0% CI for Exp(B) | |
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| Age | 0.052 | 0.017 | 9.547 | 1 | 0.002 | 1.054 | 1.019 | 1.089 |
| Location | −0.297 | 0.242 | 1.505 | 1 | 0.220 | 0.743 | 0.463 | 1.194 |
| NRAS mutation | 2.133 | 0.834 | 6.532 | 1 | 0.011 | 8.437 | 1.644 | 43.297 |
| BRAF mutation | 1.691 | 0.753 | 5.051 | 1 | 0.025 | 5.427 | 1.242 | 23.723 |
| EGFR | 0.105 | 0.178 | 0.347 | 1 | 0.556 | 1.111 | 0.783 | 1.576 |
| c-MET type | −0.105 | 0.377 | 0.077 | 1 | 0.781 | 0.901 | 0.430 | 1.887 |
| KRAS mutation | 0.117 | 0.405 | 0.084 | 1 | 0.772 | 1.124 | 0.508 | 2.487 |
| Sex | 0.046 | 0.393 | 0.014 | 1 | 0.906 | 1.047 | 0.484 | 2.265 |
| Stage | 0.917 | 0.466 | 3.874 | 1 | 0.049 | 2.501 | 1.004 | 6.232 |
| Metastasis | 1.301 | 0.927 | 1.971 | 1 | 0.160 | 3.672 | 0.597 | 22.579 |
| Liver | −0.039 | 0.582 | 0.005 | 1 | 0.946 | 0.961 | 0.307 | 3.011 |
| Lung | 0.633 | 0.262 | 5.811 | 1 | 0.016 | 1.882 | 1.126 | 3.148 |
| Other | 0.223 | 0.171 | 1.702 | 1 | 0.192 | 1.249 | 0.894 | 1.746 |
| Differentiation degree | 0.246 | 0.555 | 0.197 | 1 | 0.657 | 1.279 | 0.431 | 3.800 |
Discussion
MET, a tyrosine kinase that acts as a receptor for HGF, can activate the RAS/RAF/MAPK and PTEN/PI3K/AKT pathways. Somatic mutation and overexpression of the c-MET gene has been revealed in colorectal tumors and are related to increased tumor cell motility and invasiveness. Furthermore, new predictive and prognostic factors for the response to anti-EGFR therapy in patients with wild-type RAS tumors are actively being sought (13,14). In fact, recent studies have suggested that the abnormal activation of the MET gene may be a novel mechanism of cetuximab resistance in CRC (7,8). However, the relationship between the expression of c-MET and the clinicopathological characteristics and follow up data in CRC remains unclear, especially whether MET activation has an association with the gene profiles of KRAS, NRAS, and BRAF in patients undergoing anti-EGFR therapy.
Our study demonstrates that high protein expression of c-MET detected by immunohistochemistry was significantly related to female gender and perineural invasion of the tumor. We did not find any significant correlation between c-MET protein overexpression and tumor size, tumor location, TNM stage, and distant metastasis. The studies by Zeng et al. (15) and Liu et al. (16) did not find any association between c-MET expression and tumor size or location, consistent with the results of the present study. However, Takeuchi et al. observed that increased c-MET immunostaining was correlated with high stage (12). Zeng et al. also found c-MET gene amplification in advanced primary tumor stages. Additionally, Liu et al. showed c-MET mRNA and protein overexpression in primary CRC with advanced stage and positive nodal metastasis (16). Furthermore, Al-Maghrabi et al. reported that a high level of c-MET protein detected by immunohistochemistry was significantly associated with large tumors more than 5 cm (17). Although the same scoring system was used, their sample size was relatively smaller than in our study. This discrepancy may be attributed to the small sample size and using a different immunostaining scoring system. Here, we demonstrated that perineural invasion was significantly correlated with high c-MET protein expression of the tumor. Numerous previous studies have indicated that perineural invasion is an important negative prognostic marker in CRC, although limited reports have failed to demonstrate any prognostic effect of perineural invasion on DFS in surgically resected stage II and III CRC patients who received adjuvant treatments. As c-MET overexpression in primary CRC was significantly associated with advanced stage, positive nodal metastasis (16), and large tumor size (17), perineural invasion should be considered a high-risk feature. It could complement the classic TNM staging classification in stratifying CRC patients, especially in stages II and III, which should be treated by different treatment according to the risks. Compared with low/normal expression, c-MET overexpression significantly correlated with shorter median PFS and median OS (7), and high level EGFR expression has been reported to have an influence on the outcomes of anti-EGFR therapy in mCRC. It is reasonable to investigate the relationship between these 2 factors. Liska et al. reported that the addition of HGF to cetuximab-treated cells phosphorylated MET, but not EGFR or ErbB3, and restimulated the MAPK and AKT pathways, restored cell proliferation, and rescued cells from G1 arrest and apoptosis in CRC cells, and importantly, this effect could be abrogated by inhibiting MET activation (8). The failure for the anti-EGFR therapy for advanced mCRC is various, including the interactions between many molecular pathways. The MET amplification maybe interfere the role for EGFR-antibody in the treatment for CRC. Therefore, we evaluated the expression of EGFR into two groups, EGFR low expression (EGFR 0-+) and EGFR high expression (EGFR ++-+++), and the results revealed that c-MET protein expression had a strong correlation with the expression of EGFR, which suggests that we should pay more attention to the EGFR high expression subtype. Inhibition of the HGF-MET pathway may improve response to EGFR inhibitors in CRC, and combination therapy should be further investigated.
The activation of the EGFR signaling pathway has been reported to play an important role in the development of CRC. Currently, the advances in anti-EGFR therapy advocate that the early and comprehensive detection of KRAS, NRAS, and BRAF gene profiles is more conducive to making treatment options beneficial for CRC patients (18). In this regard, analyzing these genetic markers in different patient populations, in particular in different ethnic groups, will help determine their clinical significance. Previous evidence has shown that the mutation rate of the KRAS gene was 20–60% in CRC (19). The current study showed that the mutation frequency of the KRAS gene was higher in the cohort of CRC patients than the frequency reported worldwide (30–35%), which was 43.2%, and the mutation at codon 12 was the most common loci. NRAS mutation was detected in 3.3% (n=11) of cases, while BRAF mutation was detected in 4.7% of cases, consistent with previous reports. In addition, this study aimed to examine these genetic profiles for potential correlations with c-MET protein expression in tumor patients. The present data showed that concomitant c-MET protein overexpression in tumor tissues had no relationship with KRAS, NRAS, and BRAF genetic mutations, which has not been reported previously.
In the current study, there was no association between c-MET protein overexpression and survival, which is similar to a previous report (20). Univariate analysis showed no significant difference in the OS of patients with c-MET-high type and c-MET-low type CRC, although the median OS among patients with c-MET-high expression was slightly longer than those with c-MET-low expression. In other studies, c-MET immunostaining was found to be associated with OS (21,22). The mainstream view is that c-MET gene mutation has a certain influence on the prognosis of patients with CRC.
In summary, the results of the 337 CRC patients showed that c-MET expression was correlated with EGFR expression, female gender, and perineural invasion of the tumors. Meanwhile, concomitant c-MET protein overexpression had no relationship with KRAS, NRAS, and BRAF genetic mutations and survival. However, the relationship with the prognosis of CRC patients has not been fully elucidated.
Acknowledgments
Funding: This work was supported by a grant from the CSCO-Merck Serono Oncology Research Fund (SCORE, Y-MX2016-015), and a grant from Science and Technology Planning Project of Guangdong Province, China (2012B031800167).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-536
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-536
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-536). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Guangdong General Hospital (No. KY2020-420-01) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ma PC, Tretiakova MS, MacKinnon AC, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer 2008;47:1025-37. [Crossref] [PubMed]
- Cecchi F, Rabe DC, Bottaro DP. Targeting the HGF/Met signalling pathway in cancer. Eur J Cancer 2010;46:1260-70. [Crossref] [PubMed]
- Kammula US, Kuntz EJ, Francone TD, et al. Molecular co-expression of the c-Met oncogene and hepatocyte growth factor in primary colon cancer predicts tumor stage and clinical outcome. Cancer Lett 2007;248:219-28. [Crossref] [PubMed]
- Parizadeh SM, Jafarzadeh-Esfehani R, Fazilat-Panah D, et al. The potential therapeutic and prognostic impacts of the c-MET/HGF signaling pathway in colorectal cancer. IUBMB Life 2019;71:802-11. [Crossref] [PubMed]
- Della Corte CM, Fasano M, Papaccio F, et al. Role of HGF-MET Signaling in Primary and Acquired Resistance to Targeted Therapies in Cancer. Biomedicines 2014;2:345-58. [Crossref] [PubMed]
- Sun Y, Sun L, An Y, et al. Cabozantinib, a Novel c-Met Inhibitor, Inhibits Colorectal Cancer Development in a Xenograft Model. Med Sci Monit 2015;21:2316-21. [Crossref] [PubMed]
- Inno A, Di Salvatore M, Cenci T, et al. Is there a role for IGF1R and c-MET pathways in resistance to cetuximab in metastatic colorectal cancer? Clin Colorectal Cancer 2011;10:325-32. [Crossref] [PubMed]
- Liska D, Chen CT, Bachleitner-Hofmann T, et al. HGF rescues colorectal cancer cells from EGFR inhibition via MET activation. Clin Cancer Res 2011;17:472-82. [Crossref] [PubMed]
- Chandarlapaty S, Sawai A, Ye Q, et al. SNX2112, a synthetic heat shock protein 90 inhibitor, has potent antitumor activity against HER kinase-dependent cancers. Clin Cancer Res 2008;14:240-8. [Crossref] [PubMed]
- Kishiki T, Ohnishi H, Masaki T, et al. Overexpression of MET is a new predictive marker for anti-EGFR therapy in metastatic colorectal cancer with wild-type KRAS. Cancer Chemother Pharmacol 2014;73:749-57. [Crossref] [PubMed]
- Armstrong GR, Khot MI, Tiernan JP, et al. The utility of c-Met as a diagnostic tissue biomarker in primary colorectal cancer. Int J Exp Pathol 2021;102:172-8. [Crossref] [PubMed]
- Takeuchi H, Bilchik A, Saha S, et al. c-MET expression level in primary colon cancer: a predictor of tumor invasion and lymph node metastases. Clin Cancer Res 2003;9:1480-8. [PubMed]
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753-62. [Crossref] [PubMed]
- Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol 2010;28:1254-61. [Crossref] [PubMed]
- Zeng ZS, Weiser MR, Kuntz E, et al. c-Met gene amplification is associated with advanced stage colorectal cancer and liver metastases. Cancer Lett 2008;265:258-69. [Crossref] [PubMed]
- Liu Y, Li Q, Zhu L. Expression of the hepatocyte growth factor and c-Met in colon cancer: correlation with clinicopathological features and overall survival. Tumori 2012;98:105-12. [Crossref] [PubMed]
- Al-Maghrabi J, Emam E, Gomaa W, et al. c-MET immunostaining in colorectal carcinoma is associated with local disease recurrence. BMC Cancer 2015;15:676. [Crossref] [PubMed]
- Loupakis F, Pollina L, Stasi I, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol 2009;27:2622-9. [Crossref] [PubMed]
- Shen H, Yuan Y, Hu HG, et al. Clinical significance of K-ras and BRAF mutations in Chinese colorectal cancer patients. World J Gastroenterol 2011;17:809-16. [Crossref] [PubMed]
- Resnick MB, Routhier J, Konkin T, et al. Epidermal growth factor receptor, c-MET, beta-catenin, and p53 expression as prognostic indicators in stage II colon cancer: a tissue microarray study. Clin Cancer Res 2004;10:3069-75. [Crossref] [PubMed]
- Garouniatis A, Zizi-Sermpetzoglou A, Rizos S, et al. FAK, CD44v6, c-Met and EGFR in colorectal cancer parameters: tumour progression, metastasis, patient survival and receptor crosstalk. Int J Colorectal Dis 2013;28:9-18. [Crossref] [PubMed]
- Voutsina A, Tzardi M, Kalikaki A, et al. Combined analysis of KRAS and PIK3CA mutations, MET and PTEN expression in primary tumors and corresponding metastases in colorectal cancer. Mod Pathol 2013;26:302-13. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)



