Development and validation of a Surveillance, Epidemiology, and End Results (SEER)-based prognostic nomogram for predicting survival in elderly patients with gastric cancer after surgery
Introduction
Gastric cancer (GC) is the fifth most common malignancies worldwide with approximately 783,000 deaths in 2018, ranking third in deadly cancer (1-3). Both incidence and mortality of GC are featured by geological, diet and infection factors. Improved knowledge of prevention on Helicobacter pylori infection and related risk factors of GC have considerably reduced the overall incidence (4,5). However, increasing incidence of GC in cardia has been noticed during the past decades up to over 7-fold as many (1). Moreover, several classification systems, such as Lauren classification and an alternative one (papillary/tubular/mucinous/poorly cohesive), do not fully demonstrate clinical utility (6).
Increasing notice of heterogeneity intrigues the genomic exploration of GC, which is of essence to the full picture. The Cancer Genome Atlas (TCGA) study revealed that, comparing to classic tumor-node-metastasis (TNM) classification from WHO system, GC could be divided into four groups, including Epstein-Barr virus (EBV) positive (recurrent PIK3CA mutation and extreme DNA hypermethylation), microsatellite unstable (MSI, high mutation rates), genomically stable and chromosomal instability groups (6,7). Interestingly, TCGA indicates that genomically stable GC are more enriched at an earlier age (median age 59 years) whereas MSI tumors are found at a relatively older age (median age 72 years) (7). Another nation-wide investigation reports that age-specific trends vary significantly between older and younger groups across the United States (8). Incidence (per 100,000) drops significantly from 19.8 to 12.8 in GC patients with age 60 to 84 while increases from 0.27 to 0.45 in GC with age 25 to 39 years (8). Our previous study reported an effective prognostic nomogram for young GC (9). Only tumor size and tumor site were included for the establishment of nomogram in this study given the comparably limited sample size (276 cases as training set) (9).
Elderly GC (ELGC) remains one of the intensively investigated topics during the last decades. It is defined as GC patients over 70 years (10-12). ELGC is more correlated to intestinal type, expansive tumor growth and synchronous multiplicity with comparable poor survival outcome (11). Another study indicates that ELGC is more likely to present advanced TNM stage and larger tumor size with less odds in distant metastasis (12). Therefore, to establish a comprehensive nomogram for effective clinical practice and assessment is of significance. Noteworthy, Roberto et al. reported a prognostic nomogram for ELGC using 143 cases aged over 80 years (13). Given the shortage of large sample size in GC, it is more reliable to conduct a prognostic nomogram using public-available Surveillance, Epidemiology, and End Results (SEER) database from the National Cancer Institute (14-16). Therefore, in this study, ELGC cases from SEER database were retrieved for the establishment of nomogram. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-536).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Input data from SEER database
All the ELGC cases were retrieved from the SEER database (2004–2016) using ID 16595-Nov2019 (17). The criteria of inclusion were as follow: (I) stomach confirmed in “Site and morphology, site recode ICD-3/WHO 2008”; (II) age above 70 (including); (III) complete American Joint Committee on Cancer (AJCC) TNM information; (IV) only one primary tumor; (V) with surgery performed. Moreover, histologic type ICD-O-3 was retrieved to not only adenocarcinoma but other types of malignancies were covered, such as mixed tumor (malignant NOS). Clinical information, including age, sex and numerous variables were collected for characterization and further univariate/multivariate analysis. Total ELGC cases were randomized into training and validation groups equally. Overall survival (OS) and cancer-specific survival (CSS) were listed as primary and secondary endpoints, respectively. Moreover, x-tile program (Yale University, United States) was utilized for the optimal cutoff value of consecutive variables, including age and tumor size (18).
Statistical analysis
Univariate and multivariate analysis of all recorded clinical variables was performed. Those variables with statistically significance were extracted for the establishment of nomogram model both in OS and CSS. Next, the validation group was analyzed for assessment using the concordance index (C-index) and calibration plot. Moreover, the predict power of this nomogram was also assessed by ROC and decision curve analysis (DCA) curves. All analysis was performed by R software 3.6.0 (Vienna, Austria).
Results
Patient recruitment and characterization of cases
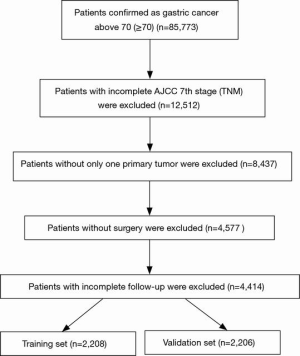
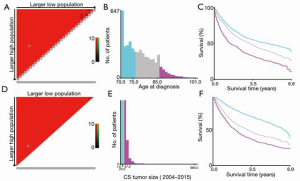
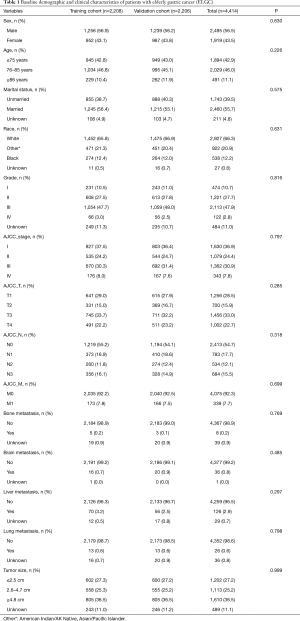
A total of 4,414 individuals with complete TNM stage and follow-up were recruited for this study, of which 2,208 were randomly in training group and 2,206 were in validation group (Figure 1). The cutoff value of age was 75 and 85 years (Figure 2A,B,C). The cutoff value of tumor size was 25 and 47 mm (Figure 2D,E,F). Among the recruited cases, 2,495 were male and 1,919 were females. Four hundred ninety-one individuals were elder than 86 years old (≥86), 1,894 were younger than 75 years old (≤75), 2,029 were between 76–85 years old. There were 1,256 individuals in T1, 700 in T2, 1,456 in T3 and 1,002 in T4. There were 2,413 individuals in N0, 783 in N1, 534 in N2 and 684 in N3. There were 4,075 individuals in distant metastasis negative and 339 in distant metastasis positive. There were only 8 positive individuals in bone metastasis, 36 in brain metastasis, 126 in liver metastasis and 26 in lung metastasis. Baseline variables were characterized between each group without significant difference (Table 1).
Full table
Establishment of nomogram models
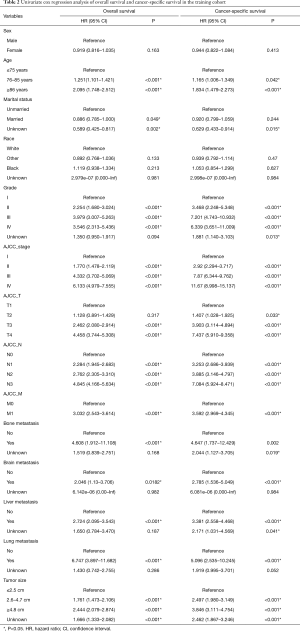
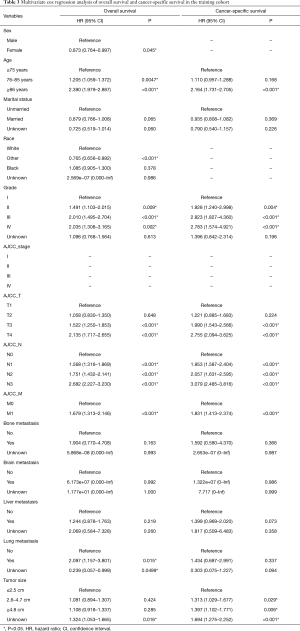
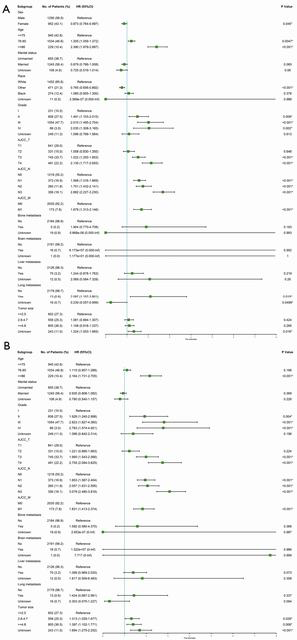
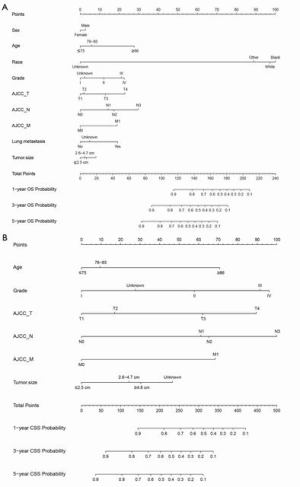
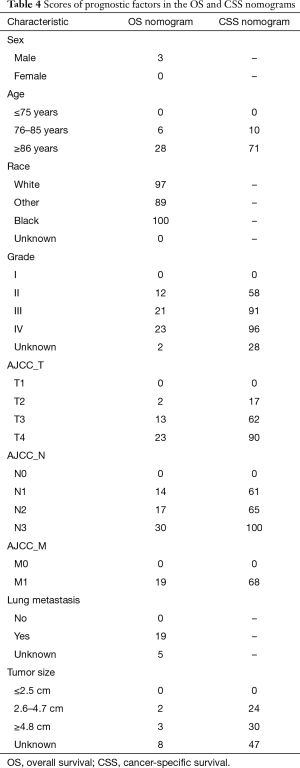
In univariate analysis of OS, significant variables (P<0.05) included age, marital status, grade, AJCC TNM stage, bone/brain/liver/lung metastasis and tumor size. In univariate analysis of CSS, significant variables (P<0.05) included age, grade, AJCC TNM stage, bone/brain/liver/lung metastasis and tumor size (Table 2). However, to fully identify potential variables that may contribute to the final outcome, variables with P<0.2 were included for multivariable analysis, separately (Table 3). In multivariate analysis of OS (Figure 3), sex, age, race, grade, TNM stage, lung metastasis and tumor size were considered as the significant variables and subjected to the establishment of nomogram (Figure 4A). In multivariable analysis of CSS (Figure 3B), age, grade, TNM, tumor size were considered as the significant variables and input to the establishment of nomogram (Figure 4B). In addition, specific scores of each included variable were displayed (Table 4).
Full table
Full table
Full table
Validation of the established nomograms
Next, to validate the power of nomograms, C-index and calibration plots were employed (Figures 5,6). Specifically, the C-index of OS in the training set was 0.730 [95% confidence interval (CI): 0.714–0.746] while in the validation set was 0.735 (95% CI: 0.719–0.751). The C-index of CSS in the training set was 0.765 (95% CI: 0.748–0.782) while in the validation set was 0.759 (95% CI: 0.742–0.776) (Table 5).
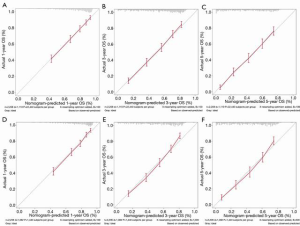
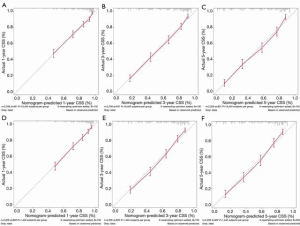

Full table
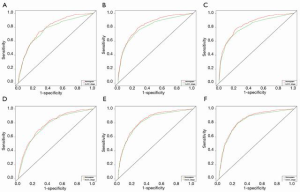
In DCA, nomograms of OS and CSS performed better than AJCC TNM stage (Figure 7). In fact, the both OS and CSS nomograms showed higher statistic power over the AJCC stage (Table 5, Figures 8,9).
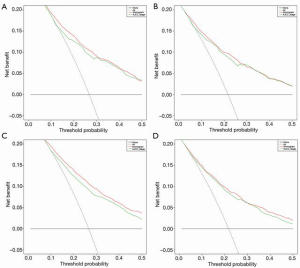
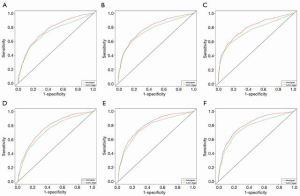
Discussion
By now, this is the first study highlighting the potential value of nomograms targeting ELGC with population dataset. For consecutive variables like age and tumor size, x-tile software was utilized for the optimal cutoff. There are a few points that should be discussed prior to conclusion.
First, although univariate analysis of OS and CSS have identified variables with significance (P<0.05), we believe a proper extension of cutoff P value (P<0.2) is reasonable and applicable. Second, sex and marital status are not included at the nomograms of OS and CSS in this study. In fact, the role of marital status in GC had been investigated (19). Qiu et al. reported that unmarried GC tended to have higher risk of cancer specific mortality (19). Our study showed no significant role of marital status associated with ELGC, indicating a possible close association of marital status to young GC rather ELGC. Third, among all four classic distant metastasis lesions, lung, brain, bone and liver, only lung metastasis was identified as a contributor to OS nomogram while none of distant metastatic lesions was associated with CSS nomogram. A similar study focusing on the nomograms of metastatic gastric adenocarcinoma with palliative gastrectomy reported that metastasis was one of the major contributors (20). However, distant lymph node metastasis and visceral metastasis were mainly evaluated. Our study complements the finding of previous study by highlighting the role of lung metastasis in the prognosis prediction of OS.
Similarly, there were numerous studies that focused on the age of GC using nomograms (21). Zhou et al. reported a prognostic nomogram to predict the OS of elderly GC using SEER data resource. However, there are several differences in between. First, the period of included cases in Zhou et al. was between 2005 and 2014 while the prognostic data of resources from our study were updated to 2016. Second, the inclusion criteria were different with only 1,445 cases in Zhou et al. results while we included 4,414 cases. The definition of ELGC was above 75 in Zhou et al. while above 70 years old in our study. In fact, we have solid evidence to support the definition (10-12). Third, both OS and CSS study were included in our study compared to Zhou et al., enhancing the distinct clinical values of the nomogram models in our results. Moreover, compared to the variables of nomogram in Zhou et al., sex, age, race, lung metastasis and tumor size also demonstrated remarkable influences on nomogram of OS, especially age itself. We believe the nomogram to predict the prognosis of ELGC opens up an opportunity to notice the significant role of age with much more subtle classification, such as patients between 70–75 years old, between 76–85 years old and patients ≥86 years old.
The limitation of this study was, however, lack of another independent clinical datasets to external validation. The predictive power of newly established nomogram required external validation and further refinement.
Conclusions
This study established and validated novel nomogram models of OS and CSS for ELGC based on population dataset.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-536
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-536). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rawla Prashanth, Barsouk Adam. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol 2019;14:26-38. [Crossref] [PubMed]
- Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol 2020;18:534-42. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Ahmed N. 23 years of the discovery of Helicobacter pylori: is the debate over? Ann Clin Microbiol Antimicrob 2005;4:17. [Crossref] [PubMed]
- Peng YC, Huang LR, Lin CL, et al. Association between proton pump inhibitors use and risk of gastric cancer in patients with GERD. Gut 2019;68:374-6. [Crossref] [PubMed]
- Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020;76:182-8. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Anderson WF, Camargo MC, Fraumeni JF Jr, et al. Age-specific trends in incidence of noncardia gastric cancer in US adults. JAMA 2010;303:1723-8. [Crossref] [PubMed]
- Yu C, Zhang Y. Development and validation of prognostic nomogram for young patients with gastric cancer. Ann Transl Med 2019;7:641. [Crossref] [PubMed]
- Saif MW, Makrilia N, Zalonis A, et al. Gastric cancer in the elderly: an overview. Eur J Surg Oncol 2010;36:709-17. [Crossref] [PubMed]
- Kitamura K, Yamaguchi T, Taniguchi H, et al. Clinicopathological characteristics of gastric cancer in the elderly. Br J Cancer 1996;73:798-802. [Crossref] [PubMed]
- Liang YX, Deng JY, Guo HH, et al. Characteristics and prognosis of gastric cancer in patients aged ≥ 70 years. World J Gastroenterol 2013;19:6568-78. [Crossref] [PubMed]
- Roberto M, Botticelli A, Strigari L, et al. Prognosis of elderly gastric cancer patients after surgery: a nomogram to predict survival. Med Oncol 2018;35:111. [Crossref] [PubMed]
- Wang SJ, Emery R, Fuller CD, et al. Conditional survival in gastric cancer: a SEER database analysis. Gastric Cancer 2007;10:153-8. [Crossref] [PubMed]
- Dikken JL, Baser RE, Gonen M, et al. Conditional probability of survival nomogram for 1-, 2-, and 3-year survivors after an R0 resection for gastric cancer. Ann Surg Oncol 2013;20:1623-30. [Crossref] [PubMed]
- Eil R, Diggs BS, Wang SJ, et al. Nomogram for predicting the benefit of neoadjuvant chemoradiotherapy for patients with esophageal cancer: A SEER-Medicare analysis. Cancer 2014;120:492-8. [Crossref] [PubMed]
- Hayat MJ, Howlader N, Reichman ME, et al. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist 2007;12:20-37. [Crossref] [PubMed]
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252-9. [Crossref] [PubMed]
- Qiu M, Yang D, Xu R. Impact of marital status on survival of gastric adenocarcinoma patients: Results from the Surveillance Epidemiology and End Results (SEER) Database. Sci Rep 2016;6:21098. [Crossref] [PubMed]
- Ma T, Wu ZJ, Xu H, et al. Nomograms for predicting survival in patients with metastatic gastric adenocarcinoma who undergo palliative gastrectomy. BMC Cancer 2019;19:852. [Crossref] [PubMed]
- Zhou X, Ding Y, Wang Y, et al. Development and Validation of a Prognostic Nomogram to Predict Overall Survival of Elderly Patients with Gastric Cancer. Research Square 2020. doi:
10.21203/rs.3.rs-36638/v1 .