
miR-4477b gene as a novel pathogenic mutation occurring during the transformation of colorectal adenoma into colorectal cancer
Introduction
A colorectal polyp is a mucosal epithelial tissue in the colon that is local bump and protrudes into the intestinal cavity. These polyps are mainly caused by the overexpression of epithelial cells and inward expansion from the mucosal surface. Pathologically, colorectal polyps are divided into tumor polyps and non-tumor polyps. The clinical symptoms of patients with non-neoplastic polyps are not obvious, and some may experience symptoms, which include slight bleeding. Generally, symptoms can improve without clinical treatment and do not transform into tumors. However, with disease development, tubular adenoma, villous adenoma, serrated adenoma, mixed adenoma, and other common tumor polyps have a high carcinogenic rate. Neoplastic polyps are regarded as having malignant potential, and colonoscopy permits most colorectal polyps to be endoscopically removed and studied pathologically (1). Early symptoms of tumor polyps are not typical and manifest mainly as abdominal distention, abdominal pain, and bloody stools; such polyps develop into colorectal cancer (CRC) after 10–20 years (2). Genetic consultation and regular examinations should be carried out. If there is any deterioration, polyps be removed as soon as possible. If treatment is not timely, CRC may develop (3).
CRC is one of the most common malignant tumors in the world. CRC ranks third and second among male and female tumors, respectively (4). At present, the occurrence of polyps and tumors caused by somatic gene mutations is a main pathogenesis of CRC (5). For example, the APC (adenomatous polyposis coli) gene is the main mutated gene in multiple abdominal polyps, especially colorectal polyps, familial polyps, and CRC (6-8). CRC evolves as a result of the stepwise accumulation of a series of genetic and epigenetic alterations in the normal colonic epithelium, leading to the development of colorectal adenomas and invasive adenocarcinomas. Based on current knowledge, the most studied epigenetic modifications in CRC include DNA methylation and histone modifications, and the role of non-coding RNAs as epigenetic regulators is also considered to be important (9).
Whole-exome sequencing (WES) has been used to detect variants in the exons of protein-coding genes, and also enables the identification of novel variants in untranslated regions and non-coding regions (10), such as non-coding RNA regulators (e.g., small RNAs, such as miRNAs and long non-coding RNAs) (11). miRNAs can affect the expression of approximately 60% of human genes, and are considered the main regulators of gene expression. Moreover, they also play important roles in physiological and pathological processes, such as cellular proliferation and development, consequently triggering diseases (12,13).
Clinical date collection showed that the brothers and sisters of the propositus had adenomas of varying degrees, and were consistent with colorectal adenomas caused by inherited genetic mutations. The clinical manifestations of siblings of the same family indicated that tumors may be caused by genetic mutations, which had been reported. Therefore, WES and peripheral blood analyses were carried out to explore the genetic mutations involved in the development of colorectal polyps into tumor polyps or even CRC, and to identify new probable genes and loci with pathogenic somatic mutations. Compared with previous studies, our detection and analysis results show that, in this family, the mutated gene was on Chr9: 68415368, which encodes miR-4477b; therefore, the gene does not code a protein. The study of this specific mutation site encoding an miRNA may deepen our understanding of the molecular mechanism of CRC metastasis, and plays an important role in predicting the relevant markers of CRC metastasis and developing key molecules as new therapeutic targets to regulate the occurrence, development, and prognosis of tumors (14). We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/jgo-20-600).
Methods
The study was approved by The Shanxi Medical University Ethics Committee (2020YX020), and informed consent was obtained from all the patients. All of the procedures involving human specimens were performed according to the Declaration of Helsinki (as revised in 2013).
Clinical data
Propositus is II:1 male, 39 years old. His main symptoms were intermittent abdominal pain and distention without regular defecation or gas for 3 months, with aggravated symptoms for 3 days (Figure 1A). He was admitted to the emergency department of the Second Hospital of Shanxi Medical University on October 31, 2018. The results of a physical examination no yellow staining of the skin and sclera of the whole body, no enlargement in the superficial lymph nodes of the whole body, coarse breathing on cardiopulmonary auscultation, no obvious dry and wet rales, abdominal distention, and positive tenderness in the middle and lower abdomen. The liver, spleen, and subcostal regions were not examined. A standing abdominal X-ray showed intestinal obstruction, and colonoscopy and pathological biopsy showed rectal adenocarcinoma (Figure 1B,C,D,E). The patient was transferred to the oncology department for 2 cycles of preoperative neoadjuvant therapy with FOLFOX6 (Oxaliplatin 150 mg; Fluorouracil 750 mg; Calcium Folinate 200 mg) treatment. Radical resection of the rectal carcinoma and left lower abdominal fistula was performed in the general surgery department of our hospital on December 10, 2018. After the operation, the patient was treated with irinotecan plus tegio (tegafur-gimeracil-oteracil potassium capsule) for 5 cycles.
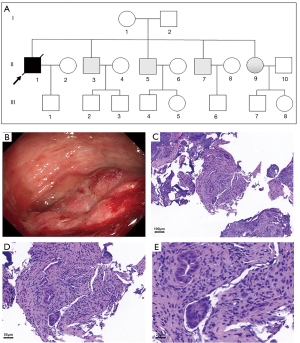
We collected 14 blood samples from the following members of each propositus’s family: propositus is II:1; eldest brother of the propositus is II:3; two sons of the eldest brother is III:2 and III:3; second brother of the propositus is II:5; son and daughter of the second brother is III:4 and III:5, respectively; third elder brother is II:5; his son is III:6; wife of the propositus is II:2; son of the propositus is III:1; and elder sister of the propositus is II:9. We also collected blood samples from the mother (I:1) and father (I:2) of the propositus.
After the propositus was diagnosed with CRC, a colonoscopy and pathological examination of the propositus’s eldest brother II:3 showed multiple colonic polyps. Histopathological results showed that the tubular adenoma was level II (Figure 2A,B,C,D,E,F,G,H). The colonoscopy and pathological results of the second brother of the propositus (II:5) showed multiple colon polyps, and the histopathological results of 1 polyp indicated tubular adenoma at level I, while the other 2 were inflammatory polyps (Figure 2I,J,K,L). The third brother of the propositus (II:7) also underwent endoscopy, and the colonoscopy results revealed sessile polyps approximately 0.5 cm × 0.5 cm in size in the ascending and descending colons (Figure 2M). Due to unknown personal reasons, these polyps were not sent to the pathology department to ascertain the pathological type. The sister of the propositus (II:9) also underwent endoscopy, and polyps of approximately 0.5 cm × 0.5 cm were found in both the transverse and sigmoid colons (Figure 2N,O). Unfortunately, because intestinal cleanliness was unsatisfactory, endoscopic polypectomy was not performed, and the pathological results are unknown.
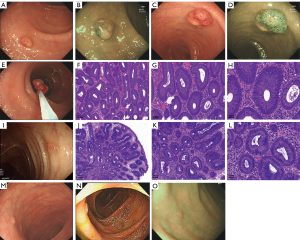
In summary, the clinical data of all of the family members showed that the propositus had colorectal adenocarcinoma. Four of the propositus’s siblings had colonic polyposis, 2 of whom were diagnosed with tubular adenomas of varying grades by endoscopy and pathology. The parents of the propositus were healthy and had no obvious clinical symptoms. The propositus’s wife and son were also used as controls for sequencing analysis.
DNA extraction
Genomic DNA (gDNA) was isolated from the peripheral blood samples of 14 family members using the iPrep PureLink gDNA Blood Kit (IS10005; Thermo Fisher Scientific, Carlsbad, CA, USA), according to the manufacturer’s instructions, and stored at –20 °C until WES and Sanger sequencing. The gDNA was sent to QIming Biotechnology (Shanghai, China) for WES of the coding organism and for DNA sequencing on the HiSeq X Ten System.
Germline sequence analysis
Fast QC software was used for quality control. The original data were compared directly to the reference genome (hg19) with Burrows-Wheeler Aligner (BWA) software (default parameters) (15), and the results were compared with Qualimap software for quality control (16). To obtain accurate results, we used GATK (16), SAMtools (17), VarScan (18), and Platypus (19) to identify mutations in the 9 samples. The 9 sample mutation files were combined into 1 mutation file, and all mutations were annotated using SnpEff software (20). Synonymous mutations in introns, intergenic regions, and exons were filtered; mutations with a population frequency >0.01 were filtered, and mutations not in line with Mendelian heritage were filtered. The mutation sites in the propositus and his brothers and sisters were identified, whereas those in his parents, wife, and controls were not. The gate gene associated with related diseases was revealed as APC, which was not mutated.
Mutation validated by polymerase chain reaction (PCR)
The full DNA sequence of the miR-4477b gene was downloaded from the National Center for Biotechnology Information website (Gene ID: 6349). PCR primers were designed using Oligo 7 software, and primers were synthesized by Shanghai Sangon Biotech. cDNA was amplified using a 2× PCR Kit (TRAN, China). The primer sequences were as follows: forward primer: 5'-TCA CCT AGC TCC CCA GAA GC-3' and reverse primer: 5'-AAC CCT CTC AAA CCC TGC TG-3'. PCR conditions were as follows: Taq was thermally activated and denatured at 94 °C for 3.5 min, and then 35 cycles were performed at 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 60 s. After gel recovery and purification, the 2-way sequencing method was used to analyze the gene sequence, which was then compared with the standard sequence.
miR-4477b target gene prediction
Two online analysis tools (TargetScan and miRDB) were used to predict the target genes of miR-4477b. To further improve the reliability of the bioinformatics analysis, Venny was used to select overlapping regions and predict the miR-4477b co-expressed target genes.
Significance analysis of target gene functions and enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) online tool was used to annotate the biologic functions of the miR-4477b target gene (6). Gene Ontology (GO) analysis includes biologic process (BP), cellular component (CC), and molecular function (MF) terms (7). GO entries with P<0.01 were selected as the enrichment functions and plotted using the R language ggplot2 package. KEGG pathway analysis was performed with the R package clusterProfiler using hypergeometric distribution for functional classification and enrichment for gene clustering (condition P<0.01) (8).
Construction of the protein-protein interaction (PPI) network and screening of hub genes
The STRING database predicts the PPI network and analyzes the functional interactions between proteins to provide insights into the genesis and development of a certain disease, with a condition confidence score the genesis and-off criterion. The MCODE plug-in of Cytoscape version 3.5.3 (http://www.cytoscape.org/download.php) was used to build the hub gene network.
Statistical analysis
In this study, BWA was used to compare the sequencing results with reference genomes, SAMtools was used to detect and filter SNP and INDEL mutations, and variant calling was used to identify mutations. Statistical methods were embedded in the software. When screening the biological functions related to the downstream target genes of miRNA, we thought that P<0.05 had statistical significance.
Results
Mutation locus analysis
According to the clinical data, the gate gene of familial adenomatous polyposis (FAP) and related gastrointestinal polyposis was APC. No mutation was found in the propositus or his brother and sister. Based on the above filter conditions, the mutation sites of the disease group and the control group were then compared. Finally, a mutation in an miRNA coding gene was detected, as shown in Figure 3. An miRNA is a length of approximately 20–24 nucleotides, and plays a variety of important regulatory roles in cells. We found that both the propositus and his brother and sister had mutations at this site, while the control did not. The mutation was validated by PCR and first-generation sequencing (Figure 3).

Target gene prediction
Prediction of miR-4477b target genes and signaling pathway analysis
TargetScan and miRanda were used to predict the target genes of miR-4477b (Figure 4A), and the intersection was examined. A total of 1,340 target genes were predicted. Furthermore, the DAVID online analysis tool was used to perform GO functional annotation on the 1,340 differentially expressed genes based on BP, CC, and MF terms. The GO functional annotation analysis of the differentially expressed genes revealed that these genes are involved mainly in the G1/S transition of the mitotic cell cycle, activation of mitogen-activated protein kinase activity, protein phosphorylation, and membrane, centrosome, cytoplasm, zinc ion-binding, protein-binding, and ligase activity (Figure 4B). KEGG pathway analysis revealed that miR-4477b regulates target genes involved mainly in the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway, regulation of the actin cytoskeleton, proteoglycans in cancer, pathways in cancer, and renal cell carcinoma (Figure 4C), which have important biologic significance.
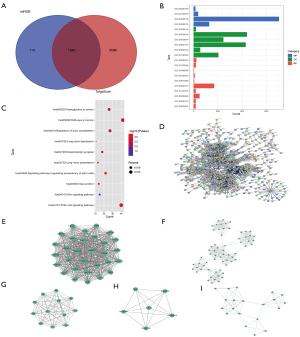
Construction of the PPI network and screening of hub genes
The STRING online database was used to preprocess the 1,340 target genes, and Cytoscape software was used to analyze the PPI network, which contains 620 nodes and 2,256 edges (Figure 4D). Five hub gene models were selected from the 1,340 target genes using the MCODE plug-in of Cytoscape (Figure 4E,F,G,H,I).
Discussion
Both adenomatous polyps and villous adenomas can predispose patients to CRC (21). In the present study, we examined a patient with CRC (the propositus) and his 3 elder brothers and elder sister, all of whom had varying degrees of colorectal polyps. Therefore, WES was performed on the fresh blood samples of all the family members. Based on the diagnostic guidelines of FAP, Lynch syndrome, and the clinical manifestations of the family, FAP and Lynch syndrome can be excluded (7,22). However, according to the comparative analysis of the whole exome/genome of this family, APC, housekeeping genes, and tumor suppressor genes associated with adenomatosis were not mutated in this family with autosomal dominant disease (6). It is unknown why some family members had colorectal polyps of varying pathological types. Therefore, in the present study, we attempted to identify other unreported gene mutations that lead to this phenomenon.
The propositus and his 4 siblings were selected as the polyp group and compared with the control group (hg19). Among the mutations revealed by WES, we observed that the gene encoding miR-4477b was mutated in the propositus and his 4 siblings. This mutation is interesting because the mutation is not in a gene that encodes a protein, but rather in a gene that encodes an miRNA. A complex regulatory mechanism might be involved in the progression from a gene mutation to adenomatous development. However, the parents of the propositus did not have similar clinical symptoms, which may indicate an acquired environmental or heterozygous mutation (23,24). The causes of the mutation will be studied in future research.
Based on the above functional and signaling pathway analyses of miR-4477b target genes, tumor-related genes are mainly involved in proteoglycans in cancer, pathways in cancer, renal cell carcinoma, and the PI3K/Akt signaling pathway. According to the existing literature, abnormalities in the PI3K signaling pathway are closely related to the occurrence and development of various tumors.
The PI3K family of lipid kinases was discovered in the mid-1980s, and has be found to play a major role in cellular functions, such as PI3K/Akt signaling pathways, which are crucial to many aspects of cancer cell growth and survival in physiological, as well as in pathological conditions (25,26). According to their structure and substrate specificity, PI3Ks belong to three distinct categories as follows: classes I, II, and III. Class I is further divided into classes IA and IB. Class IA PI3Ks seems to play the most predominant role in cancer (27,28). These kinases also heavily interact with many other pathways, including those involving hypoxia-inducible factors (25,29). The PI3K/Akt pathway is a classical pathway in cancer research and be a focus of study. Some drugs act as tumor suppressors by regulating the PI3K/Akt pathway in various cancers, including CRC (30-32). Some drugs have entered clinical trials, but some of these drugs have not worked as expected in these trials (33,34). Additional methods of influencing signaling pathways are needed. In principle and in preclinical research, miR-328-3p inhibits cell proliferation and metastasis in CRC by inhibiting the PI3K/Akt signaling pathway (35). The use of nanocomplexes loaded with miR-128-3p to enhance the effects of chemotherapy for CRC has indicated that miRNA-based cancer therapy strategies are efficient treatment methods (36). Although no classical gene was mutated during the transformation of colorectal adenoma into CRC in our study, miR-4477b may be involved in the regulation of colorectal carcinogenesis via mutation of the miR-4477b gene to reduce its copy number through the PI3K/Akt pathway.
In the clinical setting, adenomas determined by enteroscopy and pathology are likely to transform into colorectal adenocarcinomas in 10–20 years (37). This suggests that the occurrence of adenoma and CRC may have homology at the gene level (23). In addition, the propositus had CRC and his brothers and sisters had varying degrees of colorectal polyps. Therefore, gene mutations associated with hereditary colorectal polyps and CRC may occur in the exon encoding miR-4477b, and its regulatory mechanisms need to be further studied. Therefore, miR-3377b may be a new target for diagnosis or treatment.
Conclusions
Polyps may develop into CRC; however, definite mechanisms underlying the development from adenoma to CRC have not been identified. Mutations in the miR-4477b gene may represent a key mutation in colorectal carcinogenesis, and such mutations represent a multiyear cancer risk for patients and require additional attention.
Acknowledgments
We thank the study participants who helped with this research.
Funding: The study was supported by the Youth Innovation Fund of the Education Department of Shanxi Province (No. 2019L0422) and the Youth Innovation Fund of the Shanxi Province Science and Technology Department (No. 201901D211503).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/jgo-20-600
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-600
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-600). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by The Shanxi Medical University Ethics Committee (2020YX020), and informed consent was obtained from all the patients. All of the procedures involving human specimens were performed according to the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shinya H, Wolff WI. Morphology, anatomic distribution and cancer potential of colonic polyps. Ann Surg 1979;190:679-83. [Crossref] [PubMed]
- Lucci-Cordisco E, Risio M, Venesio T, et al. The growing complexity of the intestinal polyposis syndromes. Am J Med Genet A 2013;161A:2777-87. [Crossref] [PubMed]
- Lorca V, Garre P. Current status of the genetic susceptibility in attenuated adenomatous polyposis. World J Gastrointest Oncol 2019;11:1101-14. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177-93. [Crossref] [PubMed]
- Komor MA, Bosch LJ, Bounova G, et al. Consensus molecular subtype classification of colorectal adenomas. J Pathol 2018;246:266-76. [Crossref] [PubMed]
- Morin PJ. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta -catenin or APC. Science 1997;275:1787-90. [Crossref] [PubMed]
- DE Marchis ML, Tonelli F, Quaresmini D, et al. Desmoid tumors in familial adenomatous polyposis. Anticancer Res 2017;37:3357-66. [Crossref] [PubMed]
- Bahrami A, Amerizadeh F. Therapeutic potential of targeting Wnt/β-catenin pathway in treatment of colorectal cancer: Rational and progress. J Cell Biochem 2017;118:1979-83. [Crossref] [PubMed]
- Jung G, Hernández-Illán E, Moreira L, et al. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol 2020;17:111-30. [Crossref] [PubMed]
- Guo Y, Long J, He J, et al. Exome sequencing generates high quality data in non-target regions. BMC Genomics 2012;13:194. [Crossref] [PubMed]
- Warr A, Robert C, Hume D, et al. Exome sequencing: Current and future perspectives. G3 (Bethesda) 2015;5:1543-50. [Crossref] [PubMed]
- Nothnick WB. MicroRNAs and endometriosis: Distinguishing drivers from passengers in disease pathogenesis. Semin Reprod Med 2017;35:173-80. [Crossref] [PubMed]
- Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: An overview. Methods Mol Biol 2017;1509:1-10. [Crossref] [PubMed]
- de Carvalho JB, de Morais GL, Vieira T, et al. miRNA genetic variants alter their secondary structure and expression in patients with RASopathies syndromes. Front Genet 2019;10:1144. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [Crossref] [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078-9. [Crossref] [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [Crossref] [PubMed]
- Rimmer A, Phan H, Mathieson I, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 2014;46:912-8. [Crossref] [PubMed]
- Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. [Crossref] [PubMed]
- Muto T, Bussey HJR, Morson BC. The evolution of cancer of the colon and rectum. Cancer 1975;36:2251-70. [Crossref] [PubMed]
- Muller C, Matthews L, Kupfer SS, et al. Effective identification of lynch syndrome in gastroenterology practice. Curr Treat Options Gastroenterol 2019;17:666-80. [Crossref] [PubMed]
- Li J, Wang R, Zhou X, et al. Genomic and transcriptomic profiling of carcinogenesis in patients with familial adenomatous polyposis. Gut 2020;69:1283-93. [Crossref] [PubMed]
- Rashid M, Fischer A, Wilson CH, et al. Adenoma development in familial adenomatous polyposis and MUTYH-associated polyposis: Somatic landscape and driver genes. J Pathol 2016;238:98-108. [Crossref] [PubMed]
- Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol 2014;4:64. [Crossref] [PubMed]
- Whitman M, Downes CP, Keeler M, et al. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature 1988;332:644-6. [Crossref] [PubMed]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, et al. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 2010;11:329-41. [Crossref] [PubMed]
- Faes S, Dormond O. PI3K and AKT: Unfaithful partners in cancer. Int J Mol Sci 2015;16:21138-52. [Crossref] [PubMed]
- Datta SR, Brunet A, Greenberg ME. Cellular survival: A play in three Akts. Genes Dev 1999;13:2905-27. [Crossref] [PubMed]
- Ramchandani S, Naz I, Lee JH, et al. An overview of the potential antineoplastic effects of casticin. Molecules 2020;25:1287. [Crossref] [PubMed]
- Huang Z, Liu CA, Cai PZ, et al. Omega-3PUFA attenuates MNU-induced colorectal cancer in rats by blocking PI3K/AKT/Bcl-2 signaling. Onco Targets Ther 2020;13:1953-65. [Crossref] [PubMed]
- Helmy MW, Ghoneim AI, Katary MA, et al. The synergistic anti-proliferative effect of the combination of diosmin and BEZ-235 (dactolisib) on the HCT-116 colorectal cancer cell line occurs through inhibition of the PI3K/Akt/mTOR/NF-κB axis. Mol Biol Rep 2020;47:2217-30. [Crossref] [PubMed]
- Tang Y, Liu P, Tian Y, et al. Overexpression of ribonuclease inhibitor defines good prognosis and suppresses proliferation and metastasis in human colorectal cancer cells via PI3K/AKT pathway. Clin Transl Oncol 2015;17:306-13. [Crossref] [PubMed]
- Do K, Speranza G, Bishop R, et al. Biomarker-driven phase 2 study of MK-2206 and selumetinib (AZD6244, ARRY-142886) in patients with colorectal cancer. Invest New Drugs 2015;33:720-8. [Crossref] [PubMed]
- Pan S, Ren F, Li L, et al. MiR-328-3p inhibits cell proliferation and metastasis in colorectal cancer by targeting Girdin and inhibiting the PI3K/Akt signaling pathway. Exp Cell Res 2020;390:111939. [Crossref] [PubMed]
- Liu X, Dong C, Ma S, et al. Nanocomplexes loaded with miR-128-3p for enhancing chemotherapy effect of colorectal cancer through dual-targeting silence the activity of PI3K/AKT and MEK/ERK pathway. Drug Deliv 2020;27:323-33. [Crossref] [PubMed]
- Myers DJ, Arora K. Villous adenoma. StatPearls, Treasure Island, FL, 2020.
(English Language Editor: R. Scott)


