
High expression of the TEFM gene predicts poor prognosis in hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is a common primary liver malignancy worldwide (1). It is the fifth most common type of cancer worldwide and has shown a significant increase in its incidence, becoming the third leading cause of cancer-related mortality, approximately 700,000 people die of HCC every year (2). To date, liver transplantation comes the closest to cure by offering the potential for complete removal of the cancer. However, in reality, recurrence and adverse outcome rates remain high (3). In addition, HCC is usually at an advanced stage after a definite diagnosis, which delays treatment and leads to a poor prognosis (4). Therefore, in-depth exploration of the mechanism of its occurrence and development, in search of a high-sensitivity molecular marker, could greatly impact the early diagnosis, clinical treatment and prognosis of HCC.
Mitochondria are important organelles in eukaryotic cells and play an important role in the regulation of biosynthesis, cellular energy metabolism and apoptosis (5-7). Due to the essential role of mitochondria in maintaining homeostasis, mitochondrial dysfunction is associated with a variety of human diseases, such as age-dependent cardiovascular diseases, mitochondrial diabetes, neurodegenerative diseases, and cancers (8-10). Human mitochondrial transcription elongation factor (TEFM) is a key regulatory factor involved in the regulation of mitochondrial transcription and replication (11). The human TEFM gene is also named C17orf42, which is located on chromosome 17q11.2 and the full length of the complete transcript (NM_024683.3) is 1357 bp (11). The TEFM protein encodes 360 amino acid residues, and the N-terminal 1 to 35 amino acid residues are the mitochondrial targeting sequences (MTS) which help the protein enter the mitochondria (11). Recombinant TEFM protein strongly promotes mitochondrial RNA polymerase (POLRMT) processivity as it stimulates the formation of longer transcripts in vitro (12). Stimulation of POLRMT processivity by TEFM prevents the formation of G-quadraplexes that inhibit the progression of the mitochondrial elongation complex at conserved sequence block 2 (CSB2) (13,14). The capability of TEFM to abolish premature transcription termination has been proposed to function as the switch from replication to the transcription of the LSP-derived primary transcript (15). However, knockout of TEFM gene do not display an increase in mtDNA copy number. Instead, functional loss of TEFM in vivo results in an increase in premature termination, as well as a subsequent decrease in formation of replication primers (16). Moreover, functional loss of TEFM can negatively affect RNA processing in human mitochondria (17). Mitochondrial dysfunction has been found to be connected to many metabolic disorders, including cancer (18). Therefore, the putative role of TEFM in HCC was investigated since the novel therapeutic strategy is urgently needed in liver cancer. As far as we know, the expression of TEFM gene in HCC and its relationship with the progression and prognosis of patients with HCC have not been identified yet.
Based on the clinical HCC specimens and the Cancer Genome Atlas (TCGA) dataset, the purpose of the present study was to investigate the expression of TEFM gene in HCC and its correlations with the clinicopathological characteristics and prognosis of HCC patients. In this study, we want to provide the clear evidence to support the upregulation of TEFM gene expression in human HCC cells and tissues. This information would shed light on better understanding the regulatory mechanism of cancer as well as a promising therapeutic strategy for HCC. We present the study in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-120).
Methods
Patients and tissue sample
All HCC samples and matching normal liver tissues were collected at the Department of Surgery, First Affiliated Hospital of Dali University (Dali, Yunnan, China), between June 2016 and December 2018. All specimens were confirmed by the pathology department, and were stored in a −80 °C ultra-low temperature refrigerator for frozen storage 30 minutes after surgical resection. Of the 20 patients, there were 17 males and 3 females, ranging in age from 27.0 to 71.0 years, with a median age of 43.0 years. All HCC patients were diagnosed for the first time and there was no history of chemotherapy or radiotherapy before surgery. Fresh tumor tissue samples were cut from the tumor, and their corresponding adjacent normal tissues were obtained from the tumor edge ≥3 cm. All patients participating in the present study provided written informed consent, and the study was conducted in accordance with the Declaration of Helsinki (as was revised in 2013). The present study was approved by the Board and Ethical Committee of Yunnan University (ID: 2018-0201).
Cell culture
Six human HCC cell lines (SMMC-7721, BEL-7402, Hep3B, HepG2, QGY-7701, SK-Hep1), and human hepatocytes (THLE-3) were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in vitro in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Thermo Scientific, Inc., Waltham, MA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; Universal Biotech., Shanghai, China) and 1% (v/v) antibiotics (100 U/mL penicillin, 100 mg/L streptomycin; Beyotime Biotech., Shanghai, China). All cells were maintained at 37 °C in a humidified chamber with 5% CO2. All cells were authenticated via STR profiling. Mycoplasma status was checked often using the TransDetect Luciferase Mycoplasma Detection Kit (TransGen Biotech., Beijing, China).
Immunohistochemical (IHC) staining and evaluation
The specimens were fixed in 10% neutral buffered formalin, and subsequently embedded in paraffin. The paraffin-embedded tissues were sectioned at 4 µm thick and then deparaffinized with xylene and rehydrated. The slices were placed in the citric acid buffer (pH=6.0), in a microwave oven and heated for antigen retrieval for 30 min and then cooled at room temperature. Following brief proteolytic digestion and hydrogen peroxide treatment to inactivate endogenous peroxidase, the slides were incubated overnight with rabbit anti-TEFM monoclonal antibody (1:100 dilution, GeneTex, USA). After being rinsed with phosphate-buffered saline (PBS), the sections were incubated with peroxidase-conjugated secondary antibody for 2 h at room temperature. Then, the 3,3-diaminobenzidine (DAB; Zhongshan Biotech., Beijing, China) chromogenic agent was added and incubated for 15 min. The reaction was stopped by distilled water, 1% ethanol-hydrochloric acid was used for differentiation, and then the sections were washed thoroughly with distilled water. After hematoxylin counterstaining was completed, all sections were dehydrated and sealed. Cell staining was observed under an inverted microscope after routine dehydration, xylene treatment, and mounting with neutral resin. Negative controls were stained with isotype-matched control IgG. The intensity of DAB staining was analyzed using ImageJ software to assess the positive rate of tumors, and then the immunoreactivity scores (IRS) were also calculated according to our previous studies (19).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis for TEFM transcripts
Fresh HCC samples and matched normal, tumor-adjacent tissue samples (n=20) were acquired for qRT-PCR analysis from the Department of Gastroenterology at the First Affiliated Hospital of Dali University. Total RNA of HCC tissues was extracted by using the RNeasy Mini Kit (Qiagen). The first strand of cDNA synthesis from 2 µg total RNA was driven by using M-MLV reverse transcriptase (Promega). One-step qRT-PCR reactions were performed in an ABI Step One Plus Real-Time PCR System (Life Technologies) using the LightCycler FastStart DNA Master SYBR Green I Kit (Roche Diagnostics, Branchburg, New Jersey, USA). The primers for TEFM (NM_024683.4) were as follows: forward primer 5'-TGCATGTGTTGAATACAGC-3' and reverse primer 5'-AAGTTTTCTCTGTGCTCTACG-3'. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; NM_002046.7) served as an internal control gene for the quantification of TEFM mRNA levels in tissues (forward primer 5'-CGTGCCGCCTGGAGAA-3' and reverse primer 5'-CCCTCAGATGCCTGCTTCAC-3'). Reverse transcription was achieved by an incubation for 50 min at 42 °C. HieffTM Taq DNA polymerase (Yeasen Biotech., Shanghai, China) was activated by a subsequent incubation for 3 min at 94 °C. Finally, 30 amplification cycles at 94 °C for 30 seconds and 58 °C for 30 seconds, and elongation at 72 °C for 20 seconds were performed to generate 95-bp TEFM PCR products. Double delta Ct analysis was used to analyze qRT-PCR data. All tests were performed in triplicate.
Protein isolation and Western blotting
Protein samples were harvested from tissue lysates or cell lysates, and the concentration of total protein was determined by using the bicinchoninic acid (BCA; Thermo Scientific, USA) assay. Each sample equivalent of 50 mg total protein was separated by 12% SDS-PAGE gels and electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were blocked in 5% fat-free milk in tris buffered saline (TBS) for 1 h at room temperature, and then incubated at 4 °C overnight with rabbit anti-TEFM monoclonal antibody (1:1,000 dilution, GeneTex, USA). After washing with TBST (TBS+tween), the PVDF membrane was probed with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies for 2.5 h at 4 °C. Finally, the protein signals were detected with PierceTM ECL Plus Western Blotting Substrate (Thermo Scientific, USA) and semiquantitatively analyzed with Image LabTM 5.2.1 software (Bio-Rad Inc., USA). The relative protein expression level of TEFM is expressed as the ratio of the accumulated optical density (AOD) between the target band and the internal reference band. All tests were performed in triplicate.
Analysis of TEFM mRNA expression levels in HCC by GEPIA
The data of TEFM mRNA expression levels in noncancerous liver tissues (n=160) and HCC tissues (n=369) were queried from TCGA and Genotype-Tissue Expression (GTEx) datasets by the Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia2.cancer-pku.cn/). The TEFM mRNA expression levels between the multiple pathological stages in HCC were also analyzed by GEPIA.
Download of the HCC dataset from TCGA database
The mRNA-SeqV2 expression level (combining level 3 data from Illumina GA and HiSeq platform) for HCC patients was downloaded from TCGA public dataset (http://www.cbioportal.org/) by R3.6.3 (Cgdsr package). The methods of biospecimen procurement, total RNA isolation, and mRNA sequencing were previously described by the Cancer Genome Atlas Research Network (20). The level 3 TCGA data (lihc_tcga_rna_seq_v2_mrna) were obtained at from the TCGA database. Meanwhile, clinical information, OS data and DFS data were downloaded for analysis. The 373 HCC patients in the TCGA dataset were given a followed-up for 0 to 115 months. For the analysis of survival and follow-up, the date of surgery was used as the beginning of the follow-up period. The endpoint of the patient cohort analysis was the date of the last visit (if no death occurred) or the time to death. All patients who died of other diseases or accidents rather than HCC were excluded from the research cohort.
Screening of datasets for HCC in TCGA
The data for a total 373 HCC tissue samples containing TEFM expression data were downloaded by using R3.6.3 software, and 373 HCC cases had corresponding clinicopathological parameters and prognostic data. Screening and analysis of downloaded clinical case information was performed cases exclude cases with incomplete or incomplete pathological parameters and cases without prognostic follow-up. After case selection, 165 cases with complete clinicopathological parameters and survival data were incorporated. According to second-generation mRNA sequencing data, the mRNA expression of TEFM in HCC ranged from 47.2864 to 475.2475, with a median of 169.1630. To analyze the association between TEFM and clinicopathological parameters, HCC patients were divided into two subgroups (TEFMlow and TEFMhigh) according to the median value of TEFM expression levels. Accordingly, the mRNA expression level of TEFM was high in 82 cases and low in 83 cases. Detailed information on the clinical features is quantitatively assigned in Table 1.
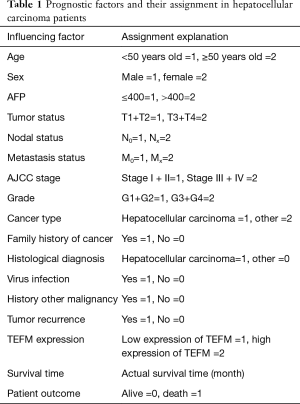
Full table
Correlation between TEFM and other mitochondrial regulatory genes or biomarker genes of HCC
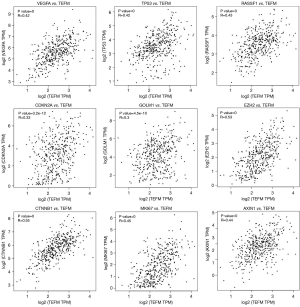
The GEPIA (http://gepia2.cancer-pku.cn/) was used to analyze the correlation between TEFM gene and other gene expression levels related to the present project from the HCC datasets in the Genotype-Tissue Expression (GTEx) projects and TCGA. The mitochondrial regulatory genes such as mitochondrial transcription termination factor 1 (MTERF1), mitochondrial transcription termination factor 2 (MTERF2), mitochondrial transcription termination factor 3 (MTERF3), mitochondrial transcription termination factor 4 (MTERF4), mitochondrial encoded NADH dehydrogenase 5 (MT-ND5), nuclear respiratory factor 1 (NRF1), mitochondrial transcription factor A (TFAM), mitochondrial RNA polymerase (POLRMT), and mitochondrial transcription factor B2 (TFB2M) were selected. After that, we searched the characteristics of HCC carcinogenesis and development (2,21) and selected HCC marker genes such as vascular endothelial growth factor (VEGF), tumor protein p53 (TP53), RAS association domain family member 1 (RASSF1), cyclin dependent kinase inhibitor 2A (CDKN2A), Golgi membrane protein 1 (GOLM1), enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), catenin beta 1 (CTNNB1), marker of proliferation Ki-67 (MIKI67), and AXIN1.
Statistical analysis
Statistical analyses were performed using the R3.6.3 software and GraphPad Prism 7.01. Categorical data are summarized as percentages and frequencies. The mRNA expression level of TEFM in the HCC dataset was evaluated with the Shapiro-Wilk method. The results showed that the TEFM mRNA expression level did not have a normal distribution. Consequently, the TEFM mRNA expression level in the HCC dataset was compared by using a Mann-Whitney U nonparametric test. Quantitative data are presented as the mean ± standard deviation (SD). A paired Student’s t-test was used to compare continuous variables in two groups. Pearson’s chi-squared test or Fisher’s exact test was used for categorical variables. The Kaplan-Meier method was used to construct OS and DFS curves, and the survival differences were assessed using the log-rank test. Moreover, univariate analyses and multivariate analyses of survival were performed to determine risk factors for HCC using the Cox proportional hazards regression model. The relative risks of death were expressed as an adjusted hazard ratio (HR) and the corresponding 95% confidence interval (CI). Pearson’s correlation test was used to analyze the association of the TEFM mRNA expression level with that of other mitochondrial regulatory genes and biomarker genes of HCC. A P value <0.05 was considered to be statistically significant.
Results
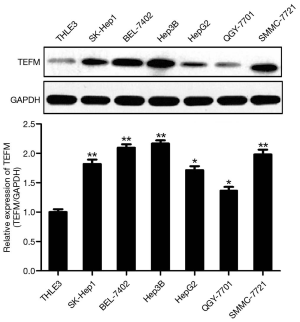
High expression of TEFM protein in six human HCC cells
Western blotting results demonstrated that the protein expression of TEFM was lowest in BEL-7402 cells and highest in Hep3B cells, and the protein expression of TEFM was significantly upregulated in six human HCC cell lines, including SMMC-7721, BEL-7402, Hep3B, HepG2, QGY-7701 and SK-Hep1, compared with human hepatocytes (THLE-3) cultured in vitro (Figure 1).
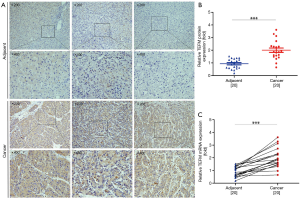
High expression of TEFM mRNA and protein in human HCC tissues
TEFM protein expression and subcellular localization in 20 HCC and 20 adjacent noncancerous liver tissues were detected by immunohistochemistry. Immunostaining of TEFM protein was primarily localized in the cytoplasm of HCC cells. Strong TEFM staining was detected in HCC tissues, whereas weak or negative staining was detected in adjacent noncancerous liver tissues. Compared with noncancerous liver tissues from the same patient, HCC tissues exhibited highly increased expression levels of TEFM protein (Figure 2A). For semi-quantification of TEFM protein levels in HCC tissues, Western blotting was conducted with tissue extracts from patient samples. Western blotting results showed that the protein expression level of TEFM in 20 HCC tissues was significantly higher than that in adjacent noncancerous liver tissues from the same patients (P<0.01) (Figure 2B). Finally, qRT-PCR results also demonstrated that the mRNA expression level of TEFM in HCC tissues was significantly increased compared with adjacent noncancerous liver tissues (P<0.01) (Figure 2C).
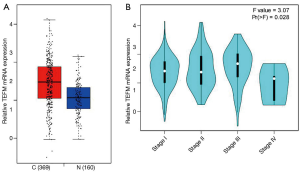
The mRNA expression of TEFM is significantly upregulated in human HCC tissues
We evaluated the mRNA expression level of TEFM in the HCC tissues (n=369) and noncancerous liver tissues (n=160) by GEPIA. The mRNA expression level of TEFM was significantly upregulated in the HCC tissues compared with the noncancerous liver tissues (P<0.05) (Figure 3A). Unfortunately, although a small increase trend could be detected in multiple pathological stages of HCC, there was no significant difference between each pathological stage of HCC (P<0.05) (Figure 3B).
Elevated expression of TEFM mRNA is correlated with the clinicopathological features of HCC
To further dissect the role of TEFM in HCC carcinogenesis, we evaluated its potential correlation with demographic factors (age, sex) and clinicopathological parameters in patients with HCC. A total of 165 cases with complete clinical pathological parameters were included in the TCGA dataset. Statistical analysis suggested that a high expression level of TEFM mRNA in HCC patients was strongly correlated with three clinicopathological parameters of HCC, including patient sex (P=0.024, χ2=5.08), serum alpha-fetoprotein (AFP) level (P=0.009, χ2=6.81) and vascular invasion (P=0.044, χ2=4.06). However, a high expression level of TEFM mRNA was not correlated with patient age, tumor volume, nodal status, metastasis status, AJCC stage, grade, cancer type, family history of cancer, virus infection, tumor recurrence and history of other malignancies in HCC (P>0.05) (Table 2).
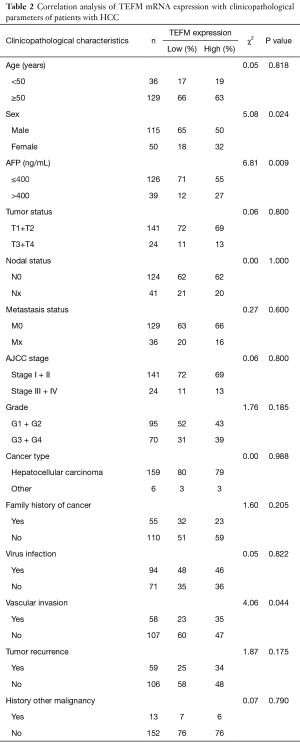
Full table
High expression of TEFM mRNA predicts poor prognosis of human HCC
To further investigate the impact of TEFM on the clinical outcome of HCC patients, Kaplan-Meier survival analysis and the log-rank test were performed based on the mRNA expression level of TEFM. Compared with patients with low expression of TEFM mRNA, patients with high expression of TEFM mRNA was significantly shortened overall survival (OS) time, poor prognosis and relapsed easily. Our results indicated that TEFM overexpression negatively correlated with OS (HR =2.01; 95% CI: 1.03–3.91, log-rank P=0.040) (Figure 4A) and DFS (HR =1.62; 95% CI: 1.01–2.66, log-rank P=0.044) (Figure 4B), which suggested that TEFM might serve as a novel prognostic marker for patients with HCC.
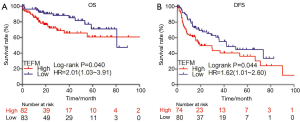
Prognostic value of clinicopathological parameters in HCC patients
To further confirm the effect of TEFM alone and among the other predictive factors of HCC prognosis, the Cox proportional hazards regression model was used for univariate and multivariate analyses to assess the clinical prognostic value of TEFM in HCC. In the univariate Cox regression analysis, age (P=0.046), tumor volume (P=0.012), vascular invasion (P=0.031) and TEFM expression (P=0.044) were prognostic factors. However, sex, serum AFP level, nodal status, metastasis status, grade, cancer type, family history of cancer, virus infection, history of other malignancies and tumor recurrence were not prognostic factors for clinical outcome (P>0.05) (Table 3). For the multivariate analysis, age (P=0.027, HR =3.11; 95% CI: 1.00–9.60), vascular invasion (P=0.023, HR =2.54; 95% CI: 1.14–5.64) and TEFM expression (P=0.041, HR =2.69; 95% CI: 1.08–3.66) were significantly independent prognostic factors in HCC (Table 3).
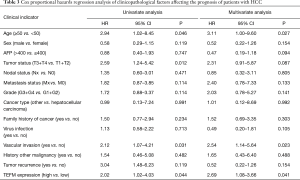
Full table
The mRNA expression level of TEFM gene is correlated with several mitochondrial regulatory genes and HCC biomarker genes
The results of gene expression correlation analysis showed that the mRNA expression level of the TEFM gene in HCC was significantly positively correlated with that of several mitochondrial transcription regulatory genes, such as MTERF1 (P=0, R=0.44), MTERF2 (P=0, R=0.50), MTERF3 (P=1e-13, R=0.35), MTERF4 (P=0, R=0.64), NRF1 (P=0, R=0.58), TFAM (P=0, R=0.70) and TFB2M (P=0, R=0.41), However, it was negatively correlated with that of the POLRMT (P=1.1e-05, R=−0.21) gene. Meanwhile, there was no correlation between TEFM and MT-ND5 gene expression levels (P>0.05) (Figure 5).
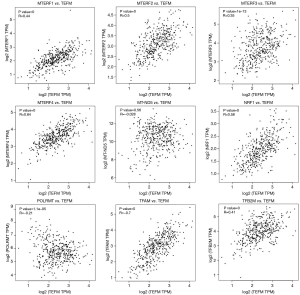
In addition, the results of gene expression correlation analysis showed that the mRNA expression level of the TEFM gene in HCC was significantly positively correlated with that of the HCC biomarker genes, such as VEGFA (P=0, R=0.42), TP53 (P=0, R=0.42), RASSF1 (P=0, R=0.43), CDKN2A (P=3.2e-12, R=0.33), GOLM1 (P=4.5e-10, R=0.30), EZH2 (P=0, R=0.53), CTNNB1 (P=0, R=0.55), MKI67 (P=0, R=0.45) and AXIN1 (P=0, R=0.44) (Figure 6).
Discussion
As a heterogeneous cancer with a complex pathogenesis, early diagnosis, early detection and early treatment have been considered as the keys to treating HCC (22,23). Therefore, it is of vital significance to identify and characterize novel biomarkers to investigate new therapeutic approaches and improve HCC patients’ clinical outcomes. Early studies have found that the occurrence of cancers is not only related to genetic material in the nucleus, but also closely related to mtDNA in the cytoplasm (24). Since the generalized acceptance of the Warburg effect as the mechanism underlying decreased oxidative phosphorylation (OXPHOS) and increased aerobic glycolytic energy production in cancer cells, a strong correlation has been found between mitochondrial dysfunction and human cancers (25,26). Studies have showed that the copy number of mtDNA varies across different tumor types, and reduced mtDNA copy number has been reported in kidney and breast cancers (27-29), as well as in HCC (30). In addition, the products of nuclear genes involved in mitochondrial transcription and replication act as communication integrators between nucleus and mitochondria, playing an essential role in both tumorigenesis and homeostase (31,32). Recent studies showed that TEFM acts as a replication-transcription switch which necessary for both replication primer formation and mtDNA transcription elongation, and knockout of TEFM gene also affects RNA processing in human mitochondria (33-35). To the best of our knowledge, the expression level of TEFM in HCC, the role of TEFM in the occurrence and development of HCC, and its impact on the prognosis of patients with HCC have not been reported yet.
In this study, we firstly investigated TEFM expression in different HCC cell lines and HCC tissues. The protein expression level of TEFM was significantly upregulated in six HCC cell lines compared with noncancerous liver cell line (THLE-3). Strong TEFM staining was detected in HCC tissues, whereas low or absent staining was shown in most noncancerous liver tissues, which indicates that TEFM may play a critical role in the carcinogenesis and progression of HCC. qRT-PCR and Western blotting results demonstrated that the mRNA and protein expression levels of TEFM in 20 HCC tissues were significantly increased compared to those in matching normal liver tissues.
In addition, we evaluated the correlations between mRNA expression level of TEFM and clinicopathological features, as well as prognostic survival in patients with HCC. Due to the relatively small number of clinical HCC samples collected in this study, R3.6.3 software was used to download the clinicopathological data of 165 HCC patients with complete parameters and TEFM mRNA expression level data from the TCGA database to confirm the validity of the results. According to the statistical analysis, a high transcriptional level of TEFM was strongly correlated with patient sex, serum AFP level and vascular invasion. Our results suggested that the increased mRNA level of TEFM was closely related to the clinicopathological features of patients with HCC. The combined use of TEFM and AFP detection could become an auxiliary means of HCC diagnosis. The Kaplan-Meier analysis and log-rank test results showed that the mRNA expression level of TEFM was significantly correlated with both OS and DFS, and high expression of TEFM mRNA predicts poor prognosis in patients with HCC. Subsequently, the Cox proportional hazards regression model was used to analyze the prognosis of patients with HCC by univariate analysis and multivariate analysis. In the multivariate analysis, age, vascular invasion and TEFM expression level were independent factors leading to poor prognosis in HCC. Finally, gene expression correlation analysis showed that the mRNA expression level of TEFM gene in HCC was significantly positively correlated with the expression levels of several mitochondrial transcription regulatory genes, such as MTERF1, MTERF2, MTERF3, MTERF4, NRF1, TFAM and TFB2M (P<0.05, R>0). However, it was significantly negatively correlated with that of the POLRMT gene (P<0.05, R<0). The results suggested that the TEFM gene may be synergistically expressed with several mitochondrial transcriptional regulatory genes and participate in the regulation of mitochondrial copy number and mRNA level in HCC, thereby affecting the type of energy metabolism in cancer cells. It is worth noting that the expression of the TEFM gene in HCC is also significantly positively correlated with the expression level of biomarker genes involved in HCC carcinogenesis. The results of gene expression correlation analysis showed that the mRNA expression level of TEFM gene in HCC was significantly positively correlated with the expression levels of the HCC biomarker genes, such as VEGFA, TP53, RASSF1, CDKN2A, GOLM1, EZH2, CTNNB1, MKI67 and AXIN1 (P<0.05, R>0). These results suggested that TEFM may be a potential diagnostic biomarker for HCC, which could strengthen HCC clinical diagnostic capabilities. Because the sensitivity and specificity of single tumor markers in clinical practice have certain limitations, combined diagnosis is one of the important methods to improve diagnostic efficiency. More research in this field is needed to investigate novel HCC biomarkers that can be used for early detection of the disease as well as new approaches to its treatment.
Despite observing the role of increased TEFM expression in HCC for the first time in our study, the possible mechanism mediated by TEFM has not yet been illuminated. Large samples of patients with long-term follow-up outcomes, and basic cellular and molecular experiments in vitro and in vivo are needed to confirm the effects of TEFM in HCC in the future. Furthermore, finding siRNA and small molecule inhibitors that effectively inhibit TEFM overexpression will be helpful to the treatment of HCC.
Conclusions
In conclusion, our data have provided clear evidence that TEFM is upregulated in human HCC cells and tissues. We also document for the first time that high expression of TEFM is associated with disease aggression and poor prognosis in patients with HCC, highlighting its potential as a prognostic biomarker and a novel molecular target for HCC treatment. Our results suggested that TEFM might be an independent predictor of clinical outcomes in HCC patients. However, further mechanistic studies on the influence of TEFM in cancer progression and prognosis are necessary to gain a full understanding of the underlying molecular effect.
Acknowledgments
Funding: This study was financially supported by the National Nature Science Foundation of China (Nos. 31160237, 31260276, 31171215, 31471187, 81560458, 31601155, 31760331), the Yunnan Provincial Science and Technology Innovation Team (2011CI123), the Reserve Talents of Young and Middle-aged Academic and Technical Leaders of Yunnan Province (No. 2017HB077), the Key Project of Yunnan Applied Basic Research Foundation (No. 202001BB050008), the Top Young Talents of Ten Thousand Talents Plan of Yunnan Province (No. 2019), and the Science and Technology Innovation Team of Dali University (ZKPY2019308, ZKLX2019320).
Footnote
Reporting Checklist: The authors present the study in accordance with the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-120
Peer Review File: Available at http://dx.doi.org/10.21037/jgo-20-120
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at: http://dx.doi.org/10.21037/jgo-20-120). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients participating in this study provided written informed consent, and the study was conducted in accordance with the Declaration of Helsinki (as was revised in 2013). This study was approved by the Board and Ethical Committee of Yunnan University (ID: 2018-0201).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol 2017;34:153-9. [Crossref] [PubMed]
- Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, et al. Hepatocellular carcinoma: From diagnosis to treatment. Surg Oncol 2016;25:74-85. [Crossref] [PubMed]
- Schlachterman A, Craft WW Jr, Hilgenfeldt E, et al. Current and future treatments for hepatocellular carcinoma. World J Gastroenterol 2015;21:8478-91. [Crossref] [PubMed]
- Zhao Y, Zhang L, Huo L, et al. Clinical significance of fucosylated GP73 in the differential diagnosis of hepatocellular carcinoma. Int J Biol Markers 2018;33:439-46. [Crossref] [PubMed]
- Wagner BK, Kitami T, Gilbert TJ, et al. Large-scale chemical dissection of mitochondrial function. Nat Biotechnol 2008;26:343-51. [Crossref] [PubMed]
- Soriano ME, Scorrano L. Traveling Bax and forth from mitochondria to control apoptosis. Cell 2011;145:15-7. [Crossref] [PubMed]
- Hengartner MO. The biochemistry of apoptosis. Nature 2000;407:770-6. [Crossref] [PubMed]
- Heller A, Brockhoff G, Goepferich A. Targeting drugs to mitochondria. Eur J Pharm Biopharm 2012;82:1-18. [Crossref] [PubMed]
- Yang Y, Karakhanova S, Hartwig W, et al. Mitochondria and mitochondrial ROS in cancer: novel targets for anticancer therapy. J Cell Physiol 2016;231:2570-81. [Crossref] [PubMed]
- Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol 2015;11:9-15. [Crossref] [PubMed]
- Minczuk M, He J, Duch AM, et al. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res 2011;39:4284-99. [Crossref] [PubMed]
- Hillen HS, Temiakov D, Cramer P. Structural basis of mitochondrial transcription. Nat Struct Mol Biol 2018;25:754-65. [Crossref] [PubMed]
- Hillen HS, Parshin AV, Agaronyan K, et al. Mechanism of transcription anti-termination in human mitochondria. Cell 2017;171:1082-93. [Crossref] [PubMed]
- Hillen HS, Morozov YI, Sarfallah A, et al. Structural basis of mitochondrial transcription initiation. Cell 2017;171:1072-81. [Crossref] [PubMed]
- Jiang S, Koolmeister C, Misic J, et al. TEFM regulates both transcription elongation and RNA processing in mitochondria. EMBO Rep 2019;20:e48101. [Crossref] [PubMed]
- Barshad G, Marom S, Cohen T, et al. Mitochondrial DNA transcription and its regulation: an evolutionary perspective. Trends Genet 2018;34:682-92. [Crossref] [PubMed]
- Posse V, Shahzad S, Falkenberg M, et al. TEFM is a potent stimulator of mitochondrial transcription elongation in vitro. Nucleic Acids Res 2015;43:2615-24. [Crossref] [PubMed]
- Cheng Z, Ristow M. Mitochondria and metabolic homeostasis. Antioxid Redox Signal 2013;19:240-2. [Crossref] [PubMed]
- Zi J, Wang W, Sun M, et al. A high expression of MTERF3 correlates with tumor progression and predicts poor outcomes in patients with brain glioma. Int J Clin Exp Pathol 2019;12:1909-20. [PubMed]
- He R, Gao L, Ma J, et al. The essential role of MTDH in the progression of HCC: a study with immunohistochemistry, TCGA, meta-analysis and in vitro investigation. Am J Transl Res 2017;9:1561-79. [PubMed]
- Banaudha KK, Verma M. Epigenetic biomarkers in liver cancer. Methods Mol Biol 2015;1238:65-76. [Crossref] [PubMed]
- Ribeiro OD, Canedo NH, Pannain VL. Immunohistochemical angiogenic biomarkers in hepatocellular carcinoma and cirrhosis: correlation with pathological features. Clinics (Sao Paulo) 2016;71:639-43. [Crossref] [PubMed]
- Lanza E, Donadon M, Poretti D, et al. Transarterial therapies for hepatocellular carcinoma. Liver Cancer 2016;6:27-33. [Crossref] [PubMed]
- Nogueira C, Almeida LS, Nesti C, et al. Syndromes associated with mitochondrial DNA depletion. Ital J Pediatr 2014;40:34. [Crossref] [PubMed]
- Ju YS, Alexandrov LB, Gerstung M, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife 2014;3:e02935. [Crossref] [PubMed]
- Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012;12:685-98. [Crossref] [PubMed]
- Shen J, Platek M, Mahasneh A, et al. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion 2010;10:62-8. [Crossref] [PubMed]
- Reznik E, Miller ML, Şenbabaoğlu Y, et al. Mitochondrial DNA copy number variation across human cancers. Elife 2016;5:e10769. [Crossref] [PubMed]
- Nilsson H, Lindgren D, Mandahl Forsberg A, et al. Primary clear cell renal carcinoma cells display minimal mitochondrial respiratory capacity resulting in pronounced sensitivity to glycolytic inhibition by 3-bromopyruvate. Cell Death Dis 2015;6:e1585. [Crossref] [PubMed]
- Qiao L, Ru G, Mao Z, et al. Mitochondrial DNA depletion, mitochondrial mutations and high TFAM expression in hepatocellular carcinoma. Oncotarget 2017;8:84373-83. [Crossref] [PubMed]
- Verma M, Naviaux RK, Tanaka M, et al. Meeting report: mitochondrial DNA and cancer epidemiology. Cancer Res 2007;67:437-9. [Crossref] [PubMed]
- Yu H, Xue C, Long M, et al. TEFM enhances transcription elongation by modifying mtRNAP pausing dynamics. Biophys J 2018;115:2295-300. [Crossref] [PubMed]
- Agaronyan K, Morozov YI, Anikin M, et al. Mitochondrial biology. replication-transcription switch in human mitochondria. Science 2015;347:548-51. [Crossref] [PubMed]
- Tan BG, Wellesley FC, Savery NJ, et al. Length heterogeneity at conserved sequence block 2 in human mitochondrial DNA acts as a rheostat for RNA polymerase POLRMT activity. Nucleic Acids Res 2016;44:7817-29. [Crossref] [PubMed]
- Li S, Wang W, Zi J, et al. Elevated expression of mitochondrial transcription elongation factor (TEFM) predicts poor prognosis in low grade glioma - an analysis of the Cancer Genome Atlas (TCGA) dataset. Transl Cancer Res 2020;9:3610-22. [Crossref]


