A cellular and bioinformatics analysis of the SENP1 SUMO isopeptidase in pancreatic cancer
Introduction
The Small Ubiquitin-related Modifier (SUMO) is a well conserved 110 amino acid protein that is post-translationally conjugated onto target proteins in a dynamic and reversible process called sumoylation. SUMO is covalently attached to target proteins through an E1, E2 and E3 enzymatic cascade. Deconjugation of SUMO from target proteins is catalyzed by sentrin-specific isopeptidases, or SENPs (1,2). A wide range of essential cellular functions are regulated by sumoylation, such as transcription, chromatin remodeling, DNA replication and cell division, among many others (3,4). Many of these essential cellular processes are misregulated in human cancers (5). As such, misregulation of SUMO conjugating and deconjugating enzymes have been implicated as contributing factors in the development and progression of many cancers (6-10). Consequently, individual pathway components have become attractive drug targets and potential biomarkers for cancer therapies (11-13). For instance, an inhibitor of the SUMO E1 conjugating enzyme has been shown to inhibit sumoylation globally and thereby decrease cancer cell proliferation and viability (8,14).
Our lab and others have identified unique roles of the SUMO isopeptidase, SENP1, in regulating genes important for cancer-related processes, such as chromosome segregation (15,16) and cellular proliferation (17-20). Through misregulation of SENP1 expression, these genes and processes can become misregulated and contribute to cancer development and progression. As such, it has been demonstrated that SENP1 expression levels can be used as a prognostic marker for a molecularly defined subset of prostate cancers (21). These promising findings prompted us to further explore reported SENP1 overexpression in pancreatic cancer (22), a lethal disease that is projected to become the second leading cause of cancer-related deaths in both men and women by 2030 (23). Lack of effective treatment options renders this disease particularly lethal and early diagnosis of the disease is essential in order to optimize treatment effectiveness and patient survival (24). The previous observation that SENP1 is up-regulated in pancreatic cancer raised the intriguing possibility that SUMO inhibitors could be used as an effective treatment option for this disease. We therefore chose to further characterize and validate SENP1 expression levels in pancreatic cancer cells and patient tissues as a step toward further establishing SENP1 as a biomarker for treatment of pancreatic cancer with a SUMO inhibitor.
We found using cell-based assays and analyses of large-scale sequencing studies from pancreatic cancer patients that in contrast to a previous report (22), SENP1 is not significantly overexpressed in pancreatic cancer. Through this work, we also provide the field with a powerful bioinformatics workflow that can be used by researchers to evaluate expression levels and genomic alterations of putative biomarkers for many common human cancers.
Materials and methods
Cell culture
We received the “normal” hTERT-transformed HPNE cell line (ATCC#: CRL-4023) graciously from Dr. Laura Wood, and three pancreatic cancer cell lines, AsPC-1 (ATCC#: CRL-1682), BxPC-3 (ATCC#: CRL-1687), CFPAC-1 (ATCC#: CRL-1918) graciously from Dr. Scott Kern. We ordered the PANC-1 cells directly from ATCC (ATCC#: CRL-1469). We also used the cervical cancer cell line, HeLa (ATCC# CCL-2), as a non-pancreas control. All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, ThermoFisher Scientific PN: 11965118) supplemented with 10% Fetal Bovine Serum (Atlanta Biologicals PN: S11150) and grown in a monolayer at 37 °C and 5% CO2. Cells were passaged one to two times per week, or until cells reached approximately 80% confluence.
All six cell lines used for this study were authenticated by the Johns Hopkins University (JHU) Genetic Resources Core Facility (GRCF) using the Promega GenePrint10 Short Tandem Repeat Profile Kit and had identities with an >80% match against the ATCC database. Additionally, we used the JHU GCRF to confirm that all six cell lines were mycoplasma free using a PCR based MycoDtect kit from Greiner Bio-One.
qRT-PCR analysis
All six cell lines were seeded at 5.0×105 cells/well in a 6-well dish and grown at 37 °C and 5% CO2 for approximately 24 hours. Total RNA was extracted using the Sigma GenElute Mammalian MiniPrep kit (Sigma PN: RTN10) following the vendor’s protocol. Extracted RNA was analyzed by nanodrop for concentration and purity. cDNA was generated using the New England BioLabs ProtoScript First Strand cDNA Synthesis Kit (NEB PN: E6300S) using 200 ng of RNA, d(T)23 VN primers and following the vendor’s recommended protocol. The qPCR reaction was performed using Bio-Rad iTaq Universal SYBR Green Supermix (Bio-Rad PN: 1725121) and following the vendors recommended protocol. qPCR runs were performed using an Applied Biosystems Quant Studio with Quant Studio v1.3.1 software. Relative SENP1 expression was calculated using average CT values from three biological replicates, a validated housekeeping gene (GAPDH), and the ΔCq equation: 2-(SENP1-GAPDH). Primer sequences:
Western blotting analysis
All six cell lines were seeded at 5.0×105 cells/well in a 6-well dish and grown at 37 °C and 5% CO2 for approximately 24 hours. Cells were harvested by scraping in 100 µL of 2X Laemmli sample buffer with 10% β-mercaptoethanol. Cells were lysed in a water bath sonicator for 3×15 second pulses, heated at 95 °C for 5 minutes, cooled and spun at 13,000 ×g for 5 minutes. Samples were loaded onto a 10-well, 10% tris-glycine gel and run at 110 V for 1 hour 15 minutes. Samples were transferred to a LF-PVDF membrane (Bio-Rad PN: 1704274) using the Bio-Rad TransBlot Turbo Mixed MW setting. Blots were blocked in 5% non-fat dry milk for 1 hour at room temperature (RT) with gentle shaking. Blots were then rinsed with 1X TBST and incubated overnight at 4 °C with a rabbit monoclonal SENP1 antibody [abcam PN: ab108981 (1:1,000)] and a mouse monoclonal alpha tubulin antibody [abcam PN: ab7291 (1:15,000)] diluted in 2% BSA, 0.02% NaN3 and 1× PBS. Blots were washed in 1xTBST and incubated in Goat anti-rabbit 800CW [LI-COR PN: 926-32211 (1:10,000)] and goat anti-mouse 680LT [LI-COR PN: 926-68020 (1:10,000)] protected from light for 1 hour at RT with gentle shaking. Blots were imaged using the LiCor Odyssey imaging system and quantitated using ImageStudio v.5.2.5 software. SENP1 signal was normalized to tubulin signal, and results were plotted using RStudio.
Immunofluorescence microscopy
Cells were seeded on coverslips at 2.5×105 cells/well in a 6-well dish and grown at 37 °C and 5% CO2 for approximately 24 hours. Media was carefully aspirated, cells were washed one time with 1× PBS, fixed in 3.5% paraformaldehyde in 1× PBS for 7 min. at RT, washed with 1× PBS and permeabilized in 0.5% Triton-X-100 in 1× PBS for 20 min. at RT. Cells were gently washed twice with 1x PBS and primary antibodies were applied [SENP1 abcam PN: ab108981 (1:500); mAb 414 abcam PN: ab24609 (1:2,000)]. Zeiss Observer Z1 fluorescence microscope with an Apotome VH optical sectioning grid was used to acquire images. SENP1 nucleoplasmic fluorescence intensity was measured using ImageJ software and graphed using RStudio.
Statistical analysis
All statistical analyses were performed using RStudio. Differences in means for qPCR and western blot data were analyzed by ANOVA, followed by a Tukey Honestly Significant Difference test with 95% confidence intervals to identify statistically significant differences between sample pairs. Differences in means for the patient data from Xena was calculated using a Students t-test. P values <0.05 were considered significant for all reported data.
Bioinformatics
Cancer Cell Line Encyclopedia (CCLE)
SENP1 mRNA expression and copy number data from human cancer cell lines was downloaded from the Broad Institute’s Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) (25). Using RStudio, data was subset and graphed to include information only from the AsPC-1, BxPC-3, CFPAC-1 and PANC-1 cell lines.
Xena
Normalized mRNA data for pancreatic cancer tissues and normal pancreas tissues were downloaded from the UCSC Xena public data hub (https://xena.ucsc.edu) (26) and opened in RStudio. The pancreatic cancer SENP1 mRNA expression values were obtained from the TCGA Pancreatic Cancer (PAAD) cohort, which had 10 studies for a total of 196 samples. The non-cancerous SENP1 mRNA expression values were downloaded from the GTEx study. In RStudio, GTEx SENP1 data was subset by organ type to include only data from normal pancreas samples, providing a total of 167 samples. Then, both TCGA and GTEx data sets were cleaned to remove samples with missing data, resulting in 178 TCGA pancreatic cancer samples, and 165 GTEx normal samples. Lastly, normality assumptions were confirmed using the Shapiro-Wilk normality test and mean expression values were compared using a Students t-test and the results were plotted using ggplot (27).
cBioPortal
The web-based cBioPortal for Cancer Genomics (28,29) (www.cbioportal.org) version 1.18.0 was used to analyze SENP1 alterations in large-scale pancreatic cancer genomic data sets. Three pancreatic adenocarcinoma cancer studies were queried: QCMG, Nature 2016 (30); TCGA, Provisional; and UTSW, Nature Communications 2015 (31). Molecular profiles were selected for Mutations and Copy Number Alterations, resulting in 751 unique patient/case sets, of which 676 were sequenced. The gene symbol “SENP1” was used to run the query. Presented data are from the OncoPrint and Cancer Types Summary tabs.
Oncomine
The Thermo Fisher Scientific Oncomine platform version 4.5 (www.oncomine.org, June 2018, Thermo Fisher Scientific, Ann Arbor, MI) was used to analyze SENP1 mRNA expression levels in pancreatic cancer patient samples as compared to adjacent normal tissues from the Badea Pancreas study (32).
GWAS Catalog
The web-based NHGRI-EBI Catalog of published genome-wide association studies (https://www.ebi.ac.uk/gwas/) was used to analyze SENP1 in GWAS studies (33).
Kaplan-Meier Plotter (KM Plot)
Relapse free and overall survival data for pancreatic cancer patients based on SENP1 mRNA expression was analyzed using the Kaplan-Meier Plotter (34) (http://kmplot.com/analysis/) using data from the pan-cancer study. The pancreatic ductal adenocarcinoma (n=177) study was selected and analysis was not restricted by subtypes.
Results
Characterizing SENP1 expression and localization in human cell lines
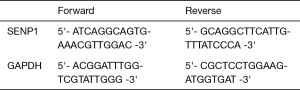
To evaluate SENP1 expression in pancreatic cancer, we first looked at SENP1 messenger RNA (mRNA) and protein levels in six human cell lines. We used the HPNE cell line, which is derived from non-cancerous pancreas tissue (35), for comparison to the pancreatic cancer cell lines AsPC-1, BxPC-3, CFPAC-1 and PANC-1, and to the cervical cancer cell line, HeLa. We found using qRT-PCR that the HPNE cells had significantly lower relative SENP1 mRNA expression as compared to all other tested cell lines (P values <0.05) (Figure 1A). Consistent with previous findings, the AsPC-1, BxPC-3 and CFPAC-1 cells had indistinguishable differences in SENP1 expression, whereas the PANC-1 cell line had the highest levels of SENP1 expression (22).
To explore whether the elevated SENP1 mRNA levels in PANC-1 cells were associated with a gene duplication event, we turned to the Broad Institute’s Cancer Cell Line Encyclopedia (CCLE, portals.broadinstitute.org/ccle) (25). We found that there were two copies of SENP1 in all four of our tested pancreatic cancer cell lines (Figure 1B), indicating that the elevated SENP1 mRNA levels in PANC-1 were not associated with a SENP1 gene duplication event. Consistent with our findings, RNA sequencing data from CCLE also showed a similar pattern of SENP1 mRNA expression for the tested pancreatic cancer cell lines.
To investigate the relationship between mRNA and protein expression levels, we probed whole cell lysates from our six human cell lines using a validated SENP1 antibody. We found that in contrast to our qPCR results, there were no significant differences in SENP1 protein levels between the tested cell lines (Figure 1C,D). Specifically, SENP1 protein levels were not as elevated in the PANC-1 cells as were expected based on our qRT-PCR results and the previous data (22). However, high molecular weight forms of SENP1 varied between PANC-1 cells and the other cell lines, as indicated by the high molecular weight bands marked by an asterisk in Figure 1C.
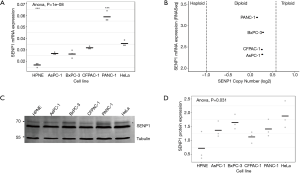
The nature of detected high molecular weight forms of SENP1 is not known, however the N-terminus of SENP1 contains multiple phosphorylation and acetylation sites, as well as predicted sumoylation and ubiquitination sites (36). Our lab and others have found that signals in the N-terminus of SENP1 determine its subcellular distribution between the nucleus, cytoplasm and nuclear pore complexes (NPCs), and it is predicted that posttranslational modifications could affect localization (15,37,38). Since we observed variations in high molecular weight forms of SENP1 in the PANC-1 cells by western blot, we investigated whether these correlate with changes in subcellular localization. We used immunofluorescence microscopy to image HPNE, AsPC-1 and PANC-1 cells co-stained for SENP1 and NPCs. Consistent with previous work from our lab (15), we found that SENP1 colocalizes with NPCs and is detectable at varying levels in small foci throughout the nucleoplasm in the three pancreas-derived cell lines (Figure 2A). More specifically, we observed similar SENP1 levels in the nucleoplasm of AsPC-1 and PANC-1 cells, and elevated levels in the nucleoplasm of HPNE cells, as quantitated in Figure 2B.
Evaluation of SENP1 in pancreatic cancer patient samples using bioinformatics
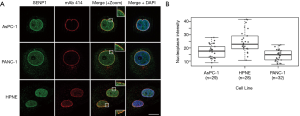
To extend our studies beyond cell lines, we next evaluated SENP1 mRNA expression, gene alterations, and survival association in pancreatic cancer patient samples, as outlined in Figure 3A. We first turned to the University of California Santa Cruz (UCSC) Xena Public Data Hub (xena.ucsc.edu) to acquire normalized mRNA data from RNA sequencing studies (26) from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression program (GTEx) (39). TCGA is a multicenter effort that profiles data at the molecular level from thousands of cancer patients across 33 cancer types. The GTEx program is another multicenter effort that generates genomic and transcriptomic profiling data from over 50 types of tissues derived from non-cancerous patient biopsies. Importantly, there are approximately equal numbers of pancreatic cancer and non-cancer samples from TCGA and GTEx, respectively, which when normalized by Xena, allows for powerful statistical comparisons between the two sources of data. To that end, we compared SENP1 mRNA expression levels from 178 pancreatic cancer samples to 165 non-cancerous pancreatic tissue samples and found that SENP1 was significantly lower in the pancreatic cancer tissues as compared to the non-cancerous tissue (P value <0.05, Figure 3B). As a second approach, we also used the Oncomine Platform by Thermo Fisher Scientific (https://www.oncomine.org) (40) as an alternative data source for evaluating SENP1 mRNA expression in pancreatic ductal adenocarcinoma tumors and matching normal pancreatic tissue samples by microarray (32). Here, we found that there is no significant difference in SENP1 mRNA expression levels between the paired tissues (Figure 3C).
To complement our SENP1 mRNA expression data, we analyzed SENP1 gene alterations using the Memorial Sloan Kettering Cancer Center (MSKCC) cBioPortal (cbioportal.org) (28,29) in 676 human pancreatic cancer samples. We found that SENP1 was amplified in 4 of the samples, deleted in 4 of the samples, and had a missense mutation of unknown significance in 1 sample (Figure 3D). Thus, the total alteration rate of the SENP1 gene in pancreatic cancer based on these samples is approximately 1.3%. Of that, only 0.6% (4/676) of the cases had a gene amplification. As a second approach, we also analyzed SENP1 single nucleotide polymorphisms (SNPs) in genome-wide association studies (GWAS) using the joint National Human Genome Research Institute (NHGRI) and European Bioinformatics Institute (EMBL-EBI) quality controlled and literature-derived catalog of published GWAS studies (https://www.ebi.ac.uk/gwas) (33). Our search identified two SENP1 variants, rs10875742 and rs2408955-T, associated with vital lung function and glycated hemoglobin levels, respectively.
Lastly, to look at the association of SENP1 expression levels and pancreatic cancer patient survival, we used the Kaplan-Meier plotter (kmplot.com) (34) to stratify patient survival data based on calculated high versus low SENP1 mRNA expression levels. The results showed no statistically significant difference in pancreatic cancer patient survival based on SENP1 mRNA expression levels (Figure 3E).
Discussion
Sumoylation regulates essential cellular processes, many of which are often misregulated in human cancers. As the SUMO pathway itself is also misregulated in numerous cancers, it has been implicated as a contributing factor in the development and progression of these diseases (4,14). Researchers have found that expression levels of individual SUMO pathway enzymes can be used as prognostic markers for cancers such as prostate and cervical cancer (13,21). Here, we used authenticated cell lines, validated reagents and data from large-scale genomics studies to evaluate the utility of SENP1 expression as a biomarker in pancreatic cancer.
To explore reported SENP1 overexpression in pancreatic cancer, we first evaluated SENP1 mRNA levels in pancreas-derived human cell lines. We found that the normal control cells had significantly lower SENP1 mRNA expression as compared to the cancer cell lines. We also found that AsPC-1, BxPC-3 and CFPAC-1 had indistinguishable differences in SENP1 expression, whereas the PANC-1 cell line had significantly higher levels of SENP1 expression. However, the magnitude of SENP1 mRNA differences between cell lines did not match previously published findings (22). In this published study, an approximate 6-fold increase in SENP1 expression in the PANC-1 cell line was observed when compared to AsPC-1 cells, whereas we observed an approximate 2-fold increase. These differences could be due to differences in cell lines (our cell line identities were validated by short tandem repeat profiling), the use of different equipment, reagents or relative expression calculations. For instance, the previous report used the ΔΔCT method, whereas we used the ΔCq method (41) since evaluating endogenous SENP1 mRNA levels does not involve the use of a treatment group. Consistent with our findings, we found that our SENP1 mRNA expression patterns were similar to those obtained by CCLE using RNA sequencing. Furthermore, data from CCLE revealed that elevated SENP1 mRNA levels in the PANC-1 cells were not associated with a gene duplication event, as all four pancreatic cancer cell lines were found to be diploid at the SENP1 locus. Surprisingly, we found that SENP1 protein levels were similar across all tested cell lines, despite higher mRNA expression in PANC-1 cells. This indicates that SENP1 protein levels are regulated post-transcriptionally, possibly at the level of translation or protein stability. Interestingly, although SENP1 protein levels did not differ between cell lines, we did observe variations in predicted modified forms of SENP1 by western blot analysis. We also observed variations in the relative distribution of SENP1 within the nucleoplasm of HPNE cells in comparison to AsPC-1 and PANC-1 cells. The prediction that these differences in observed localization reflect differences in posttranslational modifications of the SENP1 N-terminus will require future studies.
Using publicly available patient datasets, we compared SENP1 expression levels from hundreds of pancreatic cancer tissues to non-cancerous pancreas tissues. We found that SENP1 expression is lower in pancreatic cancer tissues when compared to unpaired-normal pancreas tissue, and is unchanged when compared to paired-adjacent normal pancreas tissue. The difference between these two outcomes could be explained by tissue environment, especially considering the strong desmoplastic reaction that occurs in pancreatic cancer (32). It is possible that the normal-adjacent tissues are influenced by the tumor microenvironment (TME) (42), which in turn affects SENP1 mRNA expression in the surrounding tissues. These results indicate that SENP1 levels are highest in healthy pancreas tissues and decrease in pancreatic tumor tissues. This finding is in contrast to a previous observation (22) which found elevated SENP1 mRNA levels in pancreatic ductal adenocarcinoma tissues from 22 patients as compared to adjacent normal tissues when assayed by qRT-PCR. These discordant findings could be explained by the different approaches used to evaluate SENP1 mRNA levels, the differences in sample sizes, or potential epidemiological variables related to the sources of tissue.
To further explore SENP1 in patient samples, we also looked at SENP1 gene mutations in over 600 sequenced pancreatic cancer tissues using cBioPortal. Here, we found that SENP1 was amplified in 4 of the samples, deleted in 4 of the samples, and had a missense mutation of unknown significance in 1 sample. This amounts to a 1.3% SENP1 gene alteration rate in pancreatic cancer, and furthermore, the observed differences between types of alterations suggest that SENP1 mutations in pancreatic cancer are not conserved. For comparison, KRAS, a protein well-known to promote pancreatic cancer tumorigenesis, has been found to have an alteration rate of greater than 90% and the alteration is almost always a single nucleotide variant (31).
Our search of the GWAS catalog (33) identified two SENP1 variants, rs10875742 and rs2408955-T. The rs10875742 variant was associated with vital lung function, and the rs2408955-T variant was associated with hemoglobin A1c (HbA1c) at genome-wide significance (43). Of relevance to our study, HbA1c is used to diagnose and monitor type 2 diabetes (T2D), which is a risk factor and a prognostic factor for pancreatic cancer (44). This SENP1 variant was further classified into an erythrocytic group to better define its mode of action on HbA1c, however the specific effects of this variant on the function, structure or lifespan of red blood cells have yet to be explored. Given the utility of HbA1c in diagnosing and monitoring T2D, and the link between T2D and pancreatic cancer, this variant could be of interest for further exploration.
Lastly, we found that there is no association between SENP1 expression levels and pancreatic cancer patient survival. Taken together, our data provides evidence that SENP1 is not altered at the genetic level, nor is it overexpressed in pancreatic cancer tissues or associated with patient survival. Thus, although a previous study has suggested a link between SENP1 and pancreatic cancer (22), our results do not support this finding. We therefore conclude that SENP1 is not likely to be an effective biomarker for this disease. Through this work, we have also outlined a powerful and freely-available bioinformatics workflow for the evaluation of potential biomarkers for the most common human cancers.
Conclusions
We used authenticated cell lines, validated reagents and data from large-scale genomics studies to evaluate SENP1 localization, mRNA and protein level expression, gene mutations and survival association in human pancreas cells and tissue samples. We found that SENP1 is not overexpressed in pancreatic cancer, has no association with patient survival and would therefore not make an effective biomarker. Through this work we have outlined an easy to use and freely available bioinformatics workflow for evaluating putative biomarkers for use in cancer diagnostics and therapies.
Acknowledgments
We thank Dr. Christopher Heaphy, Dr. Alan Meeker, Dr. Scott Kern, Dr. Laura Wood and Dr. Robert Anders for their discussion, guidance and reagents. Some of the results shown here are based upon data generated by the TCGA Research Network.
Funding: This work was supported by National Institute of Health (grant number GM060980 to MJ Matunis) and (grant number T32CA09110 to DM Bouchard); and was also supported in part through funding from the Sol Goldman Pancreatic Cancer Research Center.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Kunz K, Piller T, Müller S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J Cell Sci 2018. [Crossref] [PubMed]
- Zhao X. SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes. Mol Cell 2018;71:409-18. [Crossref] [PubMed]
- Eifler K, Vertegaal AC. SUMOylation-Mediated Regulation of Cell Cycle Progression and Cancer. Trends Biochem Sci 2015;40:779-93. [Crossref] [PubMed]
- Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer 2017;17:184-97. [Crossref] [PubMed]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med 2004;10:789-99. [Crossref] [PubMed]
- Bernstock JD, Ye D, Gessler FA, et al. Topotecan is a potent inhibitor of SUMOylation in glioblastoma multiforme and alters both cellular replication and metabolic programming. Sci Rep 2017;7:7425. [Crossref] [PubMed]
- Bogachek MV, Park JM, De Andrade JP, et al. Inhibiting the SUMO Pathway Represses the Cancer Stem Cell Population in Breast and Colorectal Carcinomas. Stem Cell Reports 2016;7:1140-51. [Crossref] [PubMed]
- He X, Riceberg J, Soucy T, et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat Chem Biol 2017;13:1164-71. [Crossref] [PubMed]
- Hendriks IA, D'Souza RC, Yang B, et al. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol 2014;21:927-36. [Crossref] [PubMed]
- Li R, Wei J, Jiang C, et al. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res 2013;73:5742-53. [Crossref] [PubMed]
- Cheng J, Bawa T, Lee P, et al. Role of desumoylation in the development of prostate cancer. Neoplasia 2006;8:667-76. [Crossref] [PubMed]
- Kumar A, Zhang KY. Advances in the development of SUMO specific protease (SENP) inhibitors. Comput Struct Biotechnol J 2015;13:204-11. [Crossref] [PubMed]
- Mattoscio D, Casadio C, Fumagalli M, et al. The SUMO conjugating enzyme UBC9 as a biomarker for cervical HPV infections. Ecancermedicalscience 2015;9:534. [Crossref] [PubMed]
- He X, Riceberg J, Pulukuri SM, et al. Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PLoS One 2015;10:e0123882. [Crossref] [PubMed]
- Cubeñas-Potts C, Goeres JD, Matunis MJ. SENP1 and SENP2 affect spatial and temporal control of sumoylation in mitosis. Mol Biol Cell 2013;24:3483-95. [Crossref] [PubMed]
- Lee CC, Li B, Yu H, et al. Sumoylation promotes optimal APC/C Activation and Timely Anaphase. Elife 2018. [Crossref] [PubMed]
- Wang Z, Jin J, Zhang J, et al. Depletion of SENP1 suppresses the proliferation and invasion of triple-negative breast cancer cells. Oncol Rep 2016;36:2071-8. [Crossref] [PubMed]
- Xu Y, Li J, Zuo Y, et al. SUMO-specific protease 1 regulates the in vitro and in vivo growth of colon cancer cells with the upregulated expression of CDK inhibitors. Cancer Lett 2011;309:78-84. [Crossref] [PubMed]
- Zhang QS, Zhang M, Huang XJ, et al. Downregulation of SENP1 inhibits cell proliferation, migration and promotes apoptosis in human glioma cells. Oncol Lett 2016;12:217-21. [Crossref] [PubMed]
- Zhang X, Wang H, Wang H, et al. SUMO-Specific Cysteine Protease 1 Promotes Epithelial Mesenchymal Transition of Prostate Cancer Cells via Regulating SMAD4 deSUMOylation. Int J Mol Sci 2017. [Crossref] [PubMed]
- Burdelski C, Menan D, Tsourlakis MC, et al. The prognostic value of SUMO1/Sentrin specific peptidase 1 (SENP1) in prostate cancer is limited to ERG-fusion positive tumors lacking PTEN deletion. BMC Cancer 2015;15:538. [Crossref] [PubMed]
- Ma C, Wu B, Huang X, et al. SUMO-specific protease 1 regulates pancreatic cancer cell proliferation and invasion by targeting MMP-9. Tumour Biol 2014;35:12729-35. [Crossref] [PubMed]
- Rahib L, Smith BD, Aizenberg R, et al. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res 2014;74:2913-21. [Crossref] [PubMed]
- Loosen SH, Neumann UP, Trautwein C, et al. Current and future biomarkers for pancreatic adenocarcinoma. Tumour Biol 2017;39:1010428317692231. [Crossref] [PubMed]
- Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603-7. [Crossref] [PubMed]
- Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol 2017;35:314-6. [Crossref] [PubMed]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag, 2016.
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [Crossref] [PubMed]
- Badea L, Herlea V, Dima SO, et al. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology 2008;55:2016-27. [PubMed]
- Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001-6. [Crossref] [PubMed]
- Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Lee KM, Nguyen C, Ulrich AB, et al. Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem Biophys Res Commun 2003;301:1038-44. [Crossref] [PubMed]
- Saunders B, Lyon S, Day M, et al. The Molecule Pages database. Nucleic Acids Res 2008;36:D700-6. [Crossref] [PubMed]
- Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 2012;13:755-66. [Crossref] [PubMed]
- Li SJ, Hochstrasser M. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J Cell Biol 2003;160:1069-81. [Crossref] [PubMed]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580-5. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. [Crossref] [PubMed]
- Aran D, Camarda R, Odegaard J, et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun 2017;8:1077. [Crossref] [PubMed]
- Wheeler E, Leong A, Liu CT, et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med 2017;14:e1002383. [Crossref] [PubMed]
- De Souza A, Irfan K, Masud F, et al. Diabetes Type 2 and Pancreatic Cancer: A History Unfolding. JOP 2016;17:144-8. [PubMed]