
Peptidomic changes in human serous colorectal cancer patients
Highlight box
Key findings
• Our results revealed differentially expressed peptides between the serous colorectal cancer tissue and the adjacent intestinal epithelial tissue samples.
What is known and what is new?
• There is little information for peptidomics analysis in CRC.
• We reported that 59 were significantly differentially expressed in the CRC samples and benign colonic epithelium conditions (fold changes >2, P<0.05). 25 up-regulated and 34 down-regulated peptides were detected respectively.
What is the implication, and what should change now?
• These prominently variable peptides might have an important potential role with the specific function and mechanism of them in the occurrence and development of CRC.
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and the third leading cause of cancer-related deaths, with 160,000 cases diagnosed annually in the USA (1). Surgery, radiotherapy, and chemotherapy are applied as the most common treatments for CRC (2). However, each treatment has its own indications, benefits, side effects, and potential risks. Besides, the recurrence and mortality rate after primary therapy of CRC patients is high (3). The prognosis of CRC patients may be confounded by differences in stage at presentation, tumor site, preexisting comorbidities, and type of treatment received (4). Thus, it is urgent and necessary to develop innovative therapeutic strategies for the treatment of this lethal disease. In recent years, targeted therapies, developed as promising alternative therapeutic options, have created many novel approaches for detection and treatment monitoring of CRC, such as tumor-derived exosomes (5), circulating tumor cells (6), soluble molecules (7) and miRNAs (8). For example, FOLFOX-resistance in advanced CRC is significantly associated with upregulation and downregulation of several serum miRNAs like miR-19a. However, there are still no reliable biomarkers for the diagnosis of CRC.
Peptidomics is an emerging branch of proteomics that targets endogenously produced protein fragments (9), called endogenous peptides (up to ~20 kDa), which could dynamically monitor the status of a tissue or sample. It has also been applied to screen for activators, inhibitors, proteases, and protein substrates (10). Peptides are important bioactive molecules in many physiological processes, including cancer (10,11). However, peptidomic analysis of CRC was unclear. Thus, we carried out a quantitative liquid chromatography/mass spectrometry (LC-MS) study on human serous CRC tissue and adjacent intestinal epithelial tissue to explore the relevant genes and precursor proteins. Furthermore, these different peptides between intestinal epithelial tissue and serous CRC tissue were determined and analyzed by bioinformatics. Our study aimed to identify potential antitumor peptides connected with CRC and lay a foundation for further research on possible therapeutic targets. We present the following article in accordance with the STREGA reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-188/rc).
Methods
Sample collection
We collected 3 groups of the serous CRC tissue and para-carcinoma tissue samples from the First Affiliated Hospital of Nanjing Medical University. The patients with CRC met the following inclusion criteria: (I) older than 18 years, (II) capable of giving informed consent, and (III) with serous CRC confirmed by pathology. Patients were excluded if they had any of the following: (I) other cancers; (II) a history of other cancers; (III) previous radiotherapy or chemotherapy before surgery; (IV) other diseases related to the cardiovascular system, respiratory system, genitourinary system, digestive system; or (V) acute infection. Clinical characteristics of all the patients involved are shown in Table 1. All samples were confirmed by pathology. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (No. 2018-SRFA-268). The patients were informed about the research and signed medical informed consent documents. All the tissues were collected immediately after surgery and were stored at −80 °C until use.
Table 1
| No. | Age, years | Gender | Stage |
|---|---|---|---|
| 1 | 63 | Male | T3N2bM0 |
| 2 | 63 | Male | T3N1bM0 |
| 3 | 52 | Male | T3N2AM0 |
Peptides extraction and purification
An appropriate amount of each sample was taken for extraction of this experiment. Briefly, after grinding in liquid nitrogen, samples were mixed by blowing in protein lysate. Phenylmethylsulfonyl fluoride (PMSF), 2 mM ethylenediamine tetraacetic acid (EDTA), 10 mM dithiothreitol (DTT), were added in the final solution of 1 mM concentration, which was then homogenized on ice with ultrasound and incubated on ice for 5 minutes. The mixture was centrifuged at 12,000 r/min at 4 °C for 30 minutes and the supernatant was collected into a new centrifugal tube. After being concentrated by Bradford method, an equivalent amount of protein was taken for reduction alkylation treatment. Prior to concentration measurement, equal amounts were subjected to centrifugal ultrafiltration in a 10 kd protein ultrafiltration tube (4 °C 12,000 r/min 30 min), the penetrating fluid was collected, and applied to the peptide. The polypeptide samples were desalted by C18 column, and the peptides were desalted by vacuum freeze drying.
ITRAQ labeling
The peptides were dissolved with 0.5 m triethylammonium bicarbonate (TEAB) and labeled according to the instructions of the iTRAO-8 standard kit (SCIEX, Toronto, Canada). The samples were labeled and mixed. The mixed peptide was then graded and separated using the Ultimate 3000 HPLC system (Thermo Fisher, Waltham, MA, USA). The column was Durashell-C18 (5 m, 100 A, 4.6*250 mm). Acetonitrile (ACN) concentration was gradually increased under alkaline conditions. The separation of peptides was carried out at a flow rate of 1 mL/min, and 1 tube was collected every minute. A total of 42 secondary fractions were collected and combined into 12 groups The Strata-X column was desalted and vacuum dried. The serous CRC tissue samples were labeled with reagents 116, 118, and 120, and the normal intestinal epithelial tissue samples were labeled with reagents 115, 117, and 119. The labeled peptides were frozen at –80 °C until identified analysis by liquid chromatography (LC)-tandem mass spectrometry (MS/MS).
LC-MS/MS combination analysis
The MS data was collected using the TripleTOF 5600 + liquid mass combined system (SCIEX). The polypeptide sample was dissolved in 2% CANACN with 0.1% formic acid. A TripleTOF 5600 plus mass spectrometer coupled to the Eksigent nanoLC system (SCIEX) was used for the analysis of formic acid. The polypeptide solution was added to the C18 capture column (5 m, 100 m, 20 mm) at a time gradient of 90 minutes and a flow rate of 300 µL/min at C18 Gradient elution was performed on the analysis column (3 m, 75 m, 150 mm). The 2 mobile phases were buffer A (2% ACN/0.1% formic acid/98% H2O) and buffer B (98% ACN/0.1% formic acid/2% H2O). For information dependent collection (IDA), the ion accumulation time was 250 ms. The first order mass spectra was scanned and the second order mass spectra of 30 precursor ions was collected at the ion accumulation time of 50 ms in 350–1,500 m/z. The spectrum of MS1 was collected in the range of 100 to 1,500 m/z, and the spectrum of MS2 was collected in the range of 100 to 1,500 m/z. The dynamic exclusion time was 15 seconds.
Protein identification
This experiment adopted the basic procedure of proteome identification based on MS AB Sciex 5600 and the software ProteinpilotTM V4.5 was used. Proteinpilot was used to search with all possible modification types in mind. The fault-tolerant matching function can retrieve more results than similar software under the premise of guaranteeing the reliability of identification results. For Proteinpilot in line with the results of identification, we further filtered the data and concluded that for unused score ≥1.3, the credibility level was above 95%, each protein containing at least 1 unique peptide is a credible protein, and those that did not meet the above requirements were not included in this report.
Bioinformatics analysis
The MW/PI online tool (http://web.expasy.org/compute.pi/) was applied to calculate the isoelectric point (pI) of each peptide. For further research, UniProt Database release 2015_5 (http://www.uniprot.org/), including cellular component, biological process, and molecular function, was used to perform a Gene Ontology (GO) analysis to investigate the potential functions of the peptide protein precursors. We also used Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to reveal the networks of these peptide precursors. Genes and proteins associated with differential peptides were analyzed in various respects by widely practical database including the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://string-db.org/cgi/input.pl), The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov), OncoLnc (http://www.oncolnc.org/) for protein interaction, mutated genes determination, and survival forecast, respectively. Eventually, the functional data for significantly differentiated peptides were discussed.
Statistical analysis
The 2-tailed Student’s t-test was applied for statistical analysis and P<0.05 was considered significant. Peptides with fold change (FC) >2 and P<0.05 were deemed to be a significant variation.
Results
Peptide profiling of serous CRC tissue and adjacent epithelial tissue from patients
Intracellular Peptides from the CRC and the adjacent epithelial tissue (control groups) were directly analyzed by LC-MS/MS after tandem mass tag (TMT) labeling. A total of 133 non-redundant peptides were identified by comparing 2 groups (CRC group and control group) (Table S1). Among them, 59 peptides derived from 38 precursor proteins had significantly different expression (FC >2, P<0.05), including 25 up-regulated peptides and 34 down-regulated peptides between CRC and adjacent epithelial tissues (Figure 1A). Volcano plot and heat maps were used to visualize the peptides with the highest FC (Figure 1B,1C). All of the differentially expressed peptides are listed in Table 2.
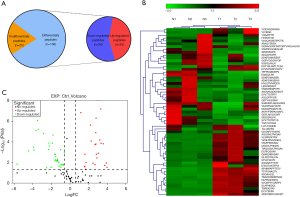
Table 2
| Gene name | Protein_ID | Full name | Peptide | Fold change | P value | Potential functiona |
|---|---|---|---|---|---|---|
| Up-regulated peptides | ||||||
| IFITM3 | Q01628 | Interferon-induced transmembrane protein 3 | APHNPAPPTSTV | 14.94 | 0.01094 | Antiviral defense, immunity, innate immunity |
| HIST1H2BC | P62807 | Histone H2B type 1-C/E/F/G/I | TSREIQTAV | 13.24 | 0.01894 | Chromosome, nucleosome core, nucleus |
| HIST2H3A | Q71DI3 | Histone H3.2 | DLRFQSSAV | 11.88 | 0.00015 | DNA-binding |
| HBB | P68871 | Hemoglobin subunit beta | VNVDEVGGEALGR | 9.91 | 0.00000 | Hypotensive agent, vasoactive |
| KRT18 | P05783 | Keratin, type I cytoskeletal 18 | SVQAPSYGARPV | 9.15 | 0.00006 | Disease mutation |
| DEFA3 | P59666 | Neutrophil defensin 3 | DCYCRIPA | 8.18 | 0.00125 | Antibiotic, antimicrobial, defensin, fungicide |
| PRDX5 | P30044 | Peroxiredoxin-5, mitochondrial | SLAPNIISQL | 7.37 | 0.00357 | Antioxidant, oxidoreductase, peroxidase |
| HIST2H2AA3 | Q6FI13 | Histone H2A type 2-A | AQGGVLPNIQAV | 6.94 | 0.0000005 | DNA-binding |
| RPS11 | P62280 | 40S ribosomal protein S11 | ADIQTERAYQKQPT | 6.87 | 0.00016 | Ribonucleoprotein, ribosomal protein, RNA-binding, rRNA-binding |
| HBA1 | P69905 | Hemoglobin subunit alpha | VLSPADKTNV | 6.45 | 0.00018 | Oxygen transport, transport |
| ALB | P02768 | Serum albumin | AASQAALGL | 6.43 | 0.01615 | Copper, lipid-binding, metal-binding, zinc |
| HIST1H1E | P10412 | Histone H1.4 | AAAGYDVEK | 5.55 | 0.01149 | DNA-binding |
| GSTP1 | P09211 | Glutathione S-transferase P | NLPINGNGKQ | 4.81 | 0.00008 | Transferase |
| RPS27L | Q71UM5 | 40S ribosomal protein S27-like | TTVFSHAQTV | 3.28 | 0.01002 | Ribonucleoprotein, ribosomal protein |
| H3F3A | P84243 | Histone H3.3 | GALQEASEAYLV | 3.03 | 0.00154 | DNA-binding |
| LTF | P02788 | Lactotransferrin | QAIAENRADAV | 2.99 | 0.02286 | Antibiotic, antimicrobial, DNA-binding, heparin-binding, Hydrolase, protease, serine protease |
| RPL37A | P61513 | 60S ribosomal protein L37a | AGGAWTYNTT | 2.98 | 0.02583 | Ribonucleoprotein, ribosomal protein |
| SNRPE | P62304 | Small nuclear ribonucleoprotein E | TLLQSVSN | 2.23 | 0.00000 | Ribonucleoprotein, RNA-binding |
| Down-regulated peptides | ||||||
| – | P0DOX7 | Immunoglobulin kappa light chain | DSTYSLSSTLTLSK | 0.60 | 0.01802 | Disulfide bond |
| IGKV2-30 | P06310 | Immunoglobulin kappa variable 2-30 | DVVMTQSPLSLPV | 0.60 | 0.01501 | Adaptive immunity, immunity |
| A2M | P01023 | Alpha-2-macroglobulin | AIGYLNTGYQR | 0.51 | 0.01666 | Protease inhibitor, serine protease inhibitor |
| HIST1H4A | P62805 | Histone H4 | LENVIRDAVT | 0.51 | 0.02907 | DNA-binding |
| ANXA2P2 | A6NMY6 | Putative annexin A2-like protein | LLYLCGGDD | 0.49 | 0.00055 | Calcium, calcium/phospholipid-binding |
| FGA | P02671 | Fibrinogen alpha chain | GEGDFLAEGGGVR | 0.44 | 0.02229 | Adaptive immunity, blood coagulation, hemostasis, immunity, innate immunity |
| HNRNPA2B1 | P22626 | Heterogeneous nuclear ribonucleoproteins A2/B1 | GGPYGGGNYGP | 0.39 | 0.00604 | Ribonucleoprotein, RNA-binding |
| VIP | P01282 | VIP peptides | NISEDPVPV | 0.39 | 0.00489 | Hormone |
| VIME | P08670 | Vimentin | ASSPGGVYATRSSAV | 0.37 | 0.00630 | Host-virus interaction |
| CFL1 | P23528 | Cofilin-1 | ASGVAVSDGVIKV | 0.36 | 0.00724 | Actin-binding |
| CNN1 | P51911 | Calponin-1 | FEPGLGMEH | 0.34 | 0.00270 | Phosphoprotein |
| PTBP1 | P26599 | Polypyrimidine tract-binding protein 1 | MDGIVPDIAVGTK | 0.33 | 0.00167 | Activator, repressor, RNA-binding |
| HNRNPR | O43390 | Heterogeneous nuclear ribonucleoprotein R | QDTYGQQWK | 0.33 | 0.00550 | Ribonucleoprotein, RNA-binding |
| RPL31 | P62899 | 60S ribosomal protein L31 | KNLQTVNVDEN | 0.33 | 0.00612 | Acetylation, phosphoprotein |
| TMSB10 | P63313 | Thymosin beta-10 | ADKPDMGEIAS | 0.32 | 0.00151 | Actin-binding |
| AHNAK | Q09666 | Neuroblast differentiation-associated protein AHNAK | ADVDISGPK | 0.21 | 0.00025 | Acetylation, isopeptide bond, methylation, phosphoprotein, Ubl conjugation |
| RTN4 | Q9NQC3 | Reticulon-4 | AAPVPTAPAAGAPL | 0.21 | 0.00013 | Neurogenesis |
| TMSB4X | P62328 | Thymosin beta-4 | SDKPDMAEIEK | 0.14 | 0.00002 | Actin-binding |
| PYY | P10082 | Peptide YY | DGPDTLLSK | 0.07 | 0.00062 | Secreted |
| KRT8 | P05787 | Keratin, type II cytoskeletal 8 | VSESSDVLPK | 0.07 | 0.00033 | Host-virus interaction |
a, data is from Uniprot; https://www.uniprot.org/.
Characterization and cleavage site analysis of the 59 differentially expressed peptides
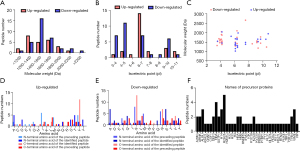
The general characteristics of the peptides were analyzed including amino acid numbers, molecular weights (MW), and pI. The MW and pI of the 59 differentially expressed peptides are shown in Figure 2A and 2B. Significantly, MWs’ range were mainly distributed between 1,200 and 2,200 Da. Meanwhile, the largest proportion of peptide pI was ranged from 3 to 10 of all peptides. The distribution of the MW relative to the pI was also investigated because of the contribution of amino acid composition and MW distribution to the specific pI distribution (Figure 2C). Given that peptidomics profiling is informative to determine protein degradation activity in various types of cancer, the cleavage sites of the differentially expressed peptides at the N- and C-terminals (C-terminal amino acid of the preceding peptide, N-terminal amino acid of the identified peptide, C-terminal amino acid of the identified peptide, and N-terminal amino acid of the subsequent peptide) were analyzed for investigation differences in 2 groups (Figure 2D,2E). In the up-regulated group, the top 4 frequency of cleavage sites contained the amino acids were valine (V), histidine (H), threonine (T), and alanine (A). In the down-regulated group, the top 4 variation cleavage sites contained the amino acids lysine (K), alanine (A), arginine (R), and serine (S). Based on the previous studies, multiple peptides may derive from the same precursor protein. Figure 2F lists the precursor proteins and demonstrates that H2A2A had the highest number of related peptides.
GO and pathway analysis of the precursor proteins of the differentially expressed peptides
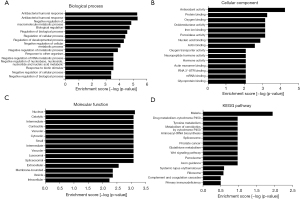
Considering the function of precursor inactivator of many peptides, we performed GO and pathway analyses to determine potential roles for these peptides based on their precursor proteins in CRC. Antibacterial humoral response, antimicrobial humoral response, negative regulation of macromolecule metabolic process, biological regulation, regulation of cellular process, and so on, were identified as the most highly enriched biological processes related to the differentially expressed peptides identified (Figure 3A). For cellular components, antioxidant activity, protein binding, oxygen binding, oxidoreductase activity, acting on peroxide as acceptor, iron ion binding, and so on, were accumulated (Figure 3B). Regarding molecular functions, nucleus, catalytic step 2 spliceosome, intermediate filament cytoskeleton, contractile fiber part, vacuolar part, cytosolic small ribosomal subunit, and so on, were the most highly abundant (Figure 3C). Besides, the KEGG pathway analysis revealed that the identified peptides were involved in malaria, drug metabolism-cytochrome P450, tyrosine metabolism, metabolism of xenobiotics by cytochrome P450, aminoacyl-tRNA biosynthesis, prostate cancer, and others (Figure 3D).
Potential function analysis of precursor proteins
Given the various differential peptides detected, 38 precursor proteins were determined eventually and there are 18 up-regulated and 20 down-regulated targets classified in Table 2. Obviously, FCs in up-regulated proteins (2.3–14.94 orders) were more drastic than in down-regulated proteins (0.07~0.6), revealing the primary variation in elevating proteins expressions. To evaluate the possible role of proteins, potential functions of 10 top variable proteins were investigated in the Web of Science database, and included 5 up-regulated proteins: IFITM3, HIST1H2BC, HIST2H3A, HBB, and KRT18 and 5 down-regulated proteins: IGKV2-30, A2M, HIST1H4A, ANXA2P2, and FGA (12-18) (Table 3). The possible roles of proteins had been identified in some previous studies, such as prognostic relevance, biomarkers, and the Notch1 pathway. Notably, some variable precursors had an unclear role in our viewpoint, which needs more detailed research in order to explain their potential mechanism and function in colon cancer.
Table 3
| Gene name | Key words of potential function | Reference | |
|---|---|---|---|
| Up-regulated | IFITM3 | Prognosis relevance | Dawei Li et al. (14), |
| HIST1H2BC | Prognosis relevance | XiaoJun Xie et al. (12) | |
| HIST2H3A | Unknown | Unknown | |
| HBB | Prognosis relevance | David S. Black et al. (13) | |
| KRT18 | Cell differentiation/Notch1 pathway | Lahdeniemi, IA et al. (15) | |
| Down-regulated | IGKV2-30 | Unknown | Unknown |
| A2M | Biomarkers & prognosis relevance | Ma Yanlei et al. (16) & Chen Jie et al. (17) | |
| HIST1H4A | Unknown | Unknown | |
| ANXA2P2 | Unknown | Unknown | |
| FGA | Diagnosis relevance/potential biomarker | Wang Hao et al. (18) |
The interaction network of peptide precursors
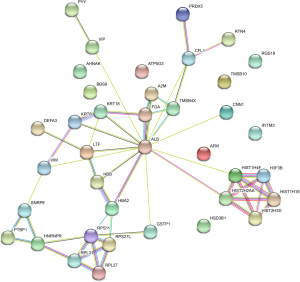
STRING analysis was widely performed to reveal the interaction network of the protein precursors of differentially expressed proteins in various cancer research (19). The interaction between 38 precursor proteins which screened before was examined for the online tool (Figure 4). In summary, there were close associations between most of predicting proteins and 2 primary groups were divided significantly. ALB, which was associated with various cancer types, plays a central role in connection with other several precursors proteins. Besides that, there are also strong connection from HIST2H2AA to other target proteins. Interestingly, although 2 studies reported this protein in bladder and lung cancer, there are limited relevant studies for its function in colon cancer (20,21). Overall, there were positively associations between proteins we identified and some proteins might have potential roles in colons cancer which remain unscreened at present, such as ALB and HIST2H2AA. However, more studies are warranted to investigate these differential proteins in the future.
Advanced genes associated peptides validation by TCGA and OncoLnc
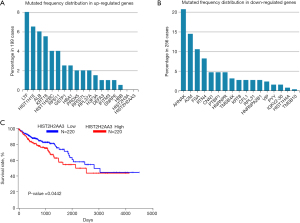
The TCGA is a publicly funded project to discover comprehensive “atlas” of cancer genomic profiles and has been application in abundant research. Based on the potential function of proteins obtained from above results, the mutated frequency and survival cure for genes corresponding all proteins (divided into up and down regulated respectively) were estimated in TCGA and OncoLnc respectively (Figure 5). In up-regulated genes, ALB had 6% mutated frequency with negative response HIST2H2AA3 in 199 cases, whereas in down-regulated genes, mutated frequency distributed from 1% (TMSB10) to above 20% (AHNAK) in 256 cases. Besides that, high expression of HIST2H2AA3 might reveal the lower survival rated (P=0.0442) estimated by OncoLnc (lower, high percentile were 50% separately). Overall, the genes-associated differential peptides might have higher mutated possibilities, which have potential function in tumorigenesis or prognosis. We hold the opinion that some genes including ALB and HIST2H2AA3 need to be explored about their potential functions in colon cancer in the future.
Discussion
Peptidomics, as an emerging branch of proteomics aimed at producing protein fragments (22), could be used to screen for amounts of disease biomarkers from serum (23), and tissues (24). The current recommended screening includes immunohistochemistry (IHC) and/or microsatelite instability (MSI) test, which gives different results for a given germline mutation sometimes (25). Evidence has displayed that significant biological differences between the CRC tissue and adjacent epithelial tissue (26,27). In recent years, an increasing number of endogenous peptides have been found to exert antitumor activity, such as the peptides derived from therapy-sensitive cancer cells that can overcome therapeutic resistance (28), and the hexapeptide PGPIPN derived from milk inhibited the metastasis of ovarian cancer cells (29). In colon cancer, the HOXB-AS3 peptide encoded by long non-coding RNA (lncRNA) HOXB-AS3 inhibited cancer growth (30). As was shown in previously mentioned literatures, peptides have been closely linked to a variety of biological processes and functions (31,32). However, the relationship between peptidomics and malignant CRC is poorly understood. Our study is the first to qualitatively compare differentially expressed peptides of CRC tissue and adjacent epithelial tissue patients. This study may pave the way for researching diagnosis and treatment markers of the CRC.
Following the experimental process, we extracted peptides fragments from CRC tissue and adjacent epithelial tissue samples by molecular weight cut off filters of 10 kDa, which could ensure the purity of the extracted peptides. In a result, 133 peptides were detected in the 6 samples, indicating that this is an effective method for extracting low-molecular-mass peptides. Functional studies of the identified peptides were conducted: 25 peptides were up-regulated and 34 peptides were down-regulated (abundance changed >2-fold) in CRC tissues compared with adjacent epithelial tissues by LC-MS/MS analysis.
Furthermore, online pI/MW tool was used to analyze the differentially expressed peptides. The results showed that the MWs of the 59 peptides were mainly distributed from 1,200 to 2,200, and the MWs of 99% of the peptides were less than 2,200, and the scope of the vast majority was 1,200 to 1,800 (79.7%). Hence, we speculated that the peptides with low MW might play a major role during the development of CRC which again highlights the effectiveness of the MWCO filter. In addition, the largest proportion of peptide pI ranged from 6 to 7, accounting for 33.9% of all peptides. In the study, we found that the most start amino acids of peptides was lysine (K), accounting for 26% in up and down regulated amino acids respectively. Lysine is an essential amino acid, necessary for human health, which seems to help the body to absorb calcium. The stoichiometry of lysine acetylation has not been explored in cancer, representing a promising field in which to increase our knowledge of how this modification is affected in cancer (33). Therefore, we suggested that the identified peptides of this study might be the key regulating factors during the CRC development progression.
Until now, the function of these different peptide precursor proteins has been poorly understood. Thus, we attempted to identify these peptidome and peptide precursors protein differentially expressed to obtain more useful information for us before the further research. Firstly, the GO analysis indicated that the molecular and cell functions of the peptides precursors mainly involved in antioxidant activity, macromolecule metabolic process, nucleus, catalytic step 2 spliceosome, which were often associated with the development and progression of tumor. Secondly, in addition, pathways (drug metabolism-cytochrome P450, tyrosine metabolism, OC signaling, metabolism of xenobiotics by cytochrome P450), participated in the occurrence of CRC. Thus, the peptides derived from these protein precursors could be putative bioactive peptides for CRC. Subsequently, we analyzed the significantly differentially expressed top 10 peptides, of which there were 5 up-regulated peptides: IFITM3, HIST1H2BC, HIST2H3A, HBB, and KRT18 and 5 down-regulated peptides: IGKV2-30, A2M, HIST1H4A, ANXA2P2, and FGA in the 54 differentially expressed peptides by the online tool SMART. Interestingly, the results showed that some have been reported through literature retrieval. IFITM3, which could be induced by interferon (IFN), is closely associated with pediatric tuberculosis in the Han Chinese Population (34,35). IFITM3 knockdown could induce cell cycle arrest in glioma cells (36). Meanwhile, Li et al. reported that IFITM3, mediated with KLF4, was overexpressed in human CRC (14). Furthermore, Fan et al. validated IFITM3 as the biomarker of early colon cancer (37). Ghosh et al. found that HBB could mediate the development of various cancers including colon cancer (38). Drew et al. forecasted that KRT18, named Keratins 18, could be considered as a molecular marker in colon adenomatous polyp and carcinoma identification (39). Interestingly, there have been few functional studies of HIST1H2BC and HIST2H3A in colon cancer. For example, Xie et al. considered this target gene a potential influencing element in prognosis of CRC (12). The role of HIST2H3A, which is considered a DNA-binding factor, has remained unclear in colon cancer to date. Based on the obvious differences identified and relevant function, we think that this gene needs more investigation in cancer research in the future. Furthermore, HIST1H4A and ANXA2P2 play an important role in DNA or phospholipid binding, suggesting their potential biological function in colon cancer.
To unravel the network of these proteins, the interaction between all targets was analyzed by online tool STRING. Obviously, ALB and HIST2H2AA (HIST2H3A) were at central location of all networks. ALB could be introduced as a possible biomarker related to colon cancer grade II to III transition (40). Given the speculation of differential gene expressions from our study, mutated frequency distribution in these genes were determined according to TCGA database to seek their variation. Notably, HIST2H2AA3 has no mutation in 199 cases which detected with higher FC rate in our study. Furthermore, the survival curve of this gene was estimated in OncoLnc for the evaluation of its potential value. Interestingly, higher expression of this gene predicted poor prognosis (P<0.05). Thus, we concluded that this gene might play an indispensable role in colon cancer, such as enhancing downstream genes expressions or according to their DNA-binding function. Based on the knowledge of persistent synthesis of HIST2H2AA3 during S-phase to package the newly replicated DNA, it might be considered as one medium for proliferation or migration in colon cancer in our review (41).
However, our study also had some shortcomings. Firstly, although we used 3 CRC tissue samples to analyze the differentially expressed peptides, the heterogeneity of the cancer population was not ruled out. The sample size was also insufficiently large. Secondly, we did not synthesize differentiated peptides for further functional verification, the function of the differentially expressed peptides in CRC progression still needs to be further investigated.
Shortly, this study screened differentially expressed peptides preliminarily based on TMT labeling combined with MS LC-MS/MS using CRC tissue samples and adjacent epithelial tissue samples from patients and has proven to be a new search for sensitive especially non-invasive tumor markers and therapeutic targets for CRC. We will also aim to validate the potential value of the above differential expression peptides as early diagnostic markers or potent therapeutic targets of CRC by expanding clinical specimens to a considerable number. Investigation is lastly required to assess the specific function and mechanism of these peptides with the CRC.
Conclusions
Shortly, this study investigated the peptides with great difference preliminarily based on TMT labeling combined with LC-MS/MS using colorectal cancer tissue samples and adjacent epithelial tissue samples from patients. Our methods have proved to be a new approach for tumor diagnostic or therapeutic markers in CRC with great sensitivity. We still need more clinical trials to validate these peptides as potential cancer biomarkers in later study with a considerable case number. Meanwhile, investigation is lastly required to assess the specific function and mechanism of them with the CRC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-188/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-188/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-188/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-188/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (No. 2018-SRFA-268). The patients were informed about the research and signed medical informed consent documents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 2009;361:2449-60. [Crossref] [PubMed]
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383:1490-502. [Crossref] [PubMed]
- Wan XB, Wang AQ, Cao J, et al. Relationships among KRAS mutation status, expression of RAS pathway signaling molecules, and clinicopathological features and prognosis of patients with colorectal cancer. World J Gastroenterol 2019;25:808-23. [Crossref] [PubMed]
- Osseis M, Nehmeh WA, Rassy N, et al. Surgery for T4 Colorectal Cancer in Older Patients: Determinants of Outcomes. J Pers Med 2022;12:1534. [Crossref] [PubMed]
- Mousavi S, Moallem R, Hassanian SM, et al. Tumor-derived exosomes: Potential biomarkers and therapeutic target in the treatment of colorectal cancer. J Cell Physiol 2019;234:12422-32. [Crossref] [PubMed]
- Grillet F, Bayet E, Villeronce O, et al. Circulating tumour cells from patients with colorectal cancer have cancer stem cell hallmarks in ex vivo culture. Gut 2017;66:1802-10. [Crossref] [PubMed]
- Kirana C, Ruszkiewicz A, Stubbs RS, et al. Soluble HLA-G is a differential prognostic marker in sequential colorectal cancer disease stages. Int J Cancer 2017;140:2577-86. [Crossref] [PubMed]
- Boussios S, Ozturk MA, Moschetta M, et al. The Developing Story of Predictive Biomarkers in Colorectal Cancer. J Pers Med 2019;9:12. [Crossref] [PubMed]
- Leichtle AB, Dufour JF, Fiedler GM. Potentials and pitfalls of clinical peptidomics and metabolomics. Swiss Med Wkly 2013;143:w13801. [Crossref] [PubMed]
- Huang X, Zhou J, Tang R, et al. Potential Significance of Peptidome in Human Ovarian Cancer for Patients With Ascites. Int J Gynecol Cancer 2018;28:355-62. [Crossref] [PubMed]
- Xu J, Wang X, Xu P, et al. Mass spectrometry-based peptidome profiling of human serous ovarian cancer tissues. Int J Biochem Cell Biol 2019;107:53-61. [Crossref] [PubMed]
- Xie XJ, Liu P, Cai CD, et al. The Generation and Validation of a 20-Genes Model Influencing the Prognosis of Colorectal Cancer. Journal of Cellular Biochemistry 2017;118:3675-85. [Crossref] [PubMed]
- Black DS, Cole SW, Christodoulou G, et al. Genomic mechanisms of fatigue in survivors of colorectal cancer. Cancer 2018;124:2637-44. [Crossref] [PubMed]
- Li D, Peng Z, Tang H, et al. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin Cancer Res 2011;17:3558-68. [Crossref] [PubMed]
- Lähdeniemi IAK, Misiorek JO, Antila CJM, et al. Keratins regulate colonic epithelial cell differentiation through the Notch1 signalling pathway. Cell Death Differ 2017;24:984-96. [Crossref] [PubMed]
- Ma Y, Zhang P, Wang F, et al. An integrated proteomics and metabolomics approach for defining oncofetal biomarkers in the colorectal cancer. Ann Surg 2012;255:720-30. [Crossref] [PubMed]
- Chen J, Zhou Y, Xu Y, et al. Low pretreatment serum globulin may predict favorable prognosis for gastric cancer patients. Tumour Biol 2016;37:3905-11. [Crossref] [PubMed]
- Wang H, Luo C, Zhu S, et al. Serum peptidome profiling for the diagnosis of colorectal cancer: discovery and validation in two independent cohorts. Oncotarget 2017;8:59376-86. [Crossref] [PubMed]
- Tang D, Zhao X, Zhang L, et al. Identification of hub genes to regulate breast cancer metastasis to brain by bioinformatics analyses. J Cell Biochem 2019;120:9522-31. [Crossref] [PubMed]
- Havaleshko DM, Cho H, Conaway M, et al. Prediction of drug combination chemosensitivity in human bladder cancer. Mol Cancer Ther 2007;6:578-86. [Crossref] [PubMed]
- Shiba-Ishii A, Kano J, Morishita Y, et al. High expression of stratifin is a universal abnormality during the course of malignant progression of early-stage lung adenocarcinoma. Int J Cancer 2011;129:2445-53. [Crossref] [PubMed]
- Latosinska A, Siwy J, Mischak H, et al. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: The past, the present, and the future. Electrophoresis 2019;40:2294-308. [Crossref] [PubMed]
- Dai Y, Hu C, Wang L, et al. Serum peptidome patterns of human systemic lupus erythematosus based on magnetic bead separation and MALDI-TOF mass spectrometry analysis. Scand J Rheumatol 2010;39:240-6. [Crossref] [PubMed]
- Chen C, Xiao D, Zhou W, et al. Comparative peptidome analyses of the profiles of the peptides ranging from 1-10 KD in CSF samples pooled from probable sporadic CJD and non-CJD patients. Prion 2012;6:46-51. [Crossref] [PubMed]
- Adeleke S, Haslam A, Choy A, et al. Microsatellite instability testing in colorectal patients with Lynch syndrome: lessons learned from a case report and how to avoid such pitfalls. Per Med 2022;19:277-86. [Crossref] [PubMed]
- Liu R, Su X, Long Y, et al. A systematic review and quantitative assessment of methylation biomarkers in fecal DNA and colorectal cancer and its precursor, colorectal adenoma. Mutat Res 2019;779:45-57. [Crossref] [PubMed]
- Jothimani G, Sriramulu S, Chabria Y, et al. A Review on Theragnostic Applications of Micrornas and Long Non- Coding RNAs in Colorectal Cancer. Curr Top Med Chem 2018;18:2614-29. [Crossref] [PubMed]
- Wang W, Gu F, Wei C, et al. PGPIPN, a therapeutic hexapeptide, suppressed human ovarian cancer growth by targeting BCL2. PLoS One 2013;8:e60701. [Crossref] [PubMed]
- Zhao M, Wei C, Yang X, et al. The milk-derived hexapeptide PGPIPN inhibits the invasion and migration of human ovarian cancer cells by regulating the expression of MTA1 and NM23H1 genes. Int J Oncol 2016;48:1721-9. [Crossref] [PubMed]
- Huang JZ, Chen M. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell 2017;68:171-84.e6. [Crossref] [PubMed]
- Neves LX, Granato DC, Busso-Lopes AF, et al. Peptidomics-Driven Strategy Reveals Peptides and Predicted Proteases Associated With Oral Cancer Prognosis. Mol Cell Proteomics 2021;20:100004. [Crossref] [PubMed]
- Foreman RE, George AL, Reimann F, et al. Peptidomics: A Review of Clinical Applications and Methodologies. Journal of Proteome Research 2021;20:3782-97. [Crossref] [PubMed]
- Gil J, Ramirez-Torres A, Encarnacion-Guevara S. Lysine acetylation and cancer: A proteomics perspective. J Proteomics 2017;150:297-309. [Crossref] [PubMed]
- Shen C, Li YJ, Yin QQ, et al. Identification of differentially expressed transcripts targeted by the knockdown of endogenous IFITM3. Mol Med Rep 2016;14:4367-73. [Crossref] [PubMed]
- Shen C, Wu XR, Jiao WW, et al. A functional promoter polymorphism of IFITM3 is associated with susceptibility to pediatric tuberculosis in Han Chinese population. PLoS One 2013;8:e67816. [Crossref] [PubMed]
- Zhao B, Wang H, Zong G, et al. The role of IFITM3 in the growth and migration of human glioma cells. BMC Neurol 2013;13:210. [Crossref] [PubMed]
- Fan J, Peng Z, Zhou C, et al. Gene-expression profiling in Chinese patients with colon cancer by coupling experimental and bioinformatic genomewide gene-expression analyses - Identification and validation of IFITM3 as a biomarker of early colon carcinogenesis. Cancer 2008;113:266-75. [Crossref] [PubMed]
- Ghosh A, De RK. Fuzzy Correlated Association Mining: Selecting altered associations among the genes, and some possible marker genes mediating certain cancers. Applied Soft Computing 2016;38:587-605. [Crossref]
- Drew JE, Farquharson AJ, Mayer CD, et al. Predictive Gene Signatures: Molecular Markers Distinguishing Colon Adenomatous Polyp and Carcinoma. Plos One 2014;9:20. [Crossref] [PubMed]
- Rostami-Nejad M, Rezaei Tavirani S, Mansouri V, et al. Gene expression profile analysis of colon cancer grade II into grade III transition by using system biology. Gastroenterol Hepatol Bed Bench 2019;12:60-6. [PubMed]
- Lyons SM, Cunningham CH, Welch JD, et al. A subset of replication-dependent histone mRNAs are expressed as polyadenylated RNAs in terminally differentiated tissues. Nucleic Acids Res 2016;44:9190-205. [Crossref] [PubMed]


