
Development and validation of an inflammatory response-related gene and clinical factor-based signature for predicting prognosis in gastric cancer
Highlight box
Key findings
• This study developed a novel prognostic model based on messenger RNA (mRNA) expression of 11 inflammatory response–related genes in gastric cancer, which was proven to be independently associated with survival outcomes in both the training and validation cohorts. The risk score was associated with immune cell infiltration and responses to chemotherapeutic, targeted, and immunotherapeutic treatments.
What is known and what is new?
• Inflammatory response is widely associated with the development of cancer and is a key consideration in cancer treatment. However, its associations with prognosis and treatment response in gastric cancer remain unclear.
• This study developed a promising inflammatory response–related prognostic model and further explored the potential roles of inflammatory response–related genes in the oncogenesis, progression, tumor immune microenvironment, and potential treatments.
What is the implication, and what should change now?
• With further investigations in a prospective setting, this inflammatory response–related model may potentially change the management of gastric cancer by enabling risk stratification and predicting response to systemic therapy.
Introduction
Gastric cancer (GC) is a commonly occurring malignant tumor, ranking fifth for incidence and fourth for mortality worldwide, with over 1 million new cases diagnosed in 2020 (1). Despite the substantial advancements in the traditional surgical techniques and chemotherapeutic regimens (i.e., 5-fluoracil–based regimen and platinum-based regimen) (2), the prognosis of patients with GC remains unsatisfactory, and thus advanced targeted therapy and immunotherapy has been applied to GC. However, only a portion of patients respond well and benefit from these treatment approaches due to tumor heterogeneity, the tumor immune microenvironment (TME), a diversity of risk factors, and varied genetic characteristics among the population (3). Given the high heterogeneity of the disease, identifying biomarkers for predicting prognosis and treatment response is urgently needed, as this can enable risk stratification and provide information for individualized treatment options, facilitating more potent adjuvant therapy and intensive follow-up visits in high-risk patients (4). The TNM staging system of the American Joint Committee on Cancer (AJCC) is widely used for GC staging (5,6). However, patients with the same AJCC stage and similar clinical treatments may have a variety of prognoses, which suggests that more parameters beyond staging are needed for the estimating prognosis in GC. In addition to tumor stage, previous studies have attempted to explore the potential of other clinicopathologic features as prognostic factors, although the predictive efficacies were unsatisfactory in GC patients, including the histologic grade (7), abnormal tumor markers (8), and lymph node invasion (9,10). Recently, next-generation sequencing (NGS)–based approaches, including RNA-sequencing (RNA-seq), whole-exome sequencing (WES), whole-genome sequencing (WGS), and targeted sequencing have provided insights into the molecular mechanisms of GC development (8,11,12). Potential tumor mutational burden (TMB) identified with advanced sequencing technologies is also a promising therapeutic and prognostic biomarker for immunotherapy and tumor management (13). Additionally, together with the establishment of public databases, the sheer volume of sequencing data has enabled numerous studies to explore the prognostic and predictive gene markers for GC management (11). For instance, based on artificial intelligence algorithms, such as least absolute shrinkage and selection operator (LASSO) (14) and principal component analysis (PCA) (15), novel prognostic signatures have been developed based on pyroptosis-related long noncoding RNAs (16) and genes (17), metastasis-related epithelial–mesenchymal transition pathways (18), angiogenesis-associated genes (19), and immune ferroptosis–related genes (14). The unsatisfactory predictive accuracies, the prognostic value validated in limited populations, and the high cost of tests owing to a large number of features involved in the model have limited their clinical applications. Despite the expanded knowledge of the molecular mechanisms related to GC, the establishment of a universally applicable robust model remains distant. Prognostic nomograms are pictorial and quantitative models with high precision and predicting capacity which have been established in clinical practice to assess cancer survival, including GC (10,20). By considering crucial prognostic indicators, nomograms can more correctly estimate survival for individual patients than the AJCC staging method (21).
Inflammatory responses are critically involved in tumor initiation, growth, progression, metastasis, and response to therapy (22-24). Several prognostic scoring systems based on hematological inflammatory parameters have been developed to predict survival outcomes in GC (25-30). For instance, the Glasgow prognostic score has been shown capable of independently predicting survival in patients with GC (31). Besides, the systemic immune-inflammatory index, which consists of lymphocytes, neutrophils, and platelets (platelets × neutrophils/lymphocytes), was identified as an effective prognostic signature in GC by various studies and meta-analysis (32), suggesting the potential inflammatory-related mechanisms are of great potential to predict the GC prognosis and treatment response accurately. Most of the previous related studies have focused on the systemic inflammatory response, with the prognostic value of the messenger RNA (mRNA) expression of the inflammatory response–related genes in cancer tissues remaining largely unknown.
In anticancer therapy, acute inflammatory response has been recognized as a strong modulator of the TME, and chronic inflammation causes an increase in immunosuppressive cell populations in the TME (33). Chemotherapeutic agents and radiation therapy can induce immunogenic cell death and activate antitumor T-cell responses (34-36). On the other hand, these therapies may induce chronic inflammation events which may cause resistance and failure of therapy (33,37). One of the major obstacles for immunotherapy is an immunosuppressive TME shaped by chronic inflammation (38), while acute inflammation induced by other therapies can improve immunotherapy’s effectiveness (39,40). Thus, the inflammatory response–related signatures of the tumor may also predict its response to treatments.
In the present study, an 11-gene prognostic model was developed in The Cancer Genome Atlas-stomach adenocarcinoma (TCGA-STAD) cohort and validated in 3 independent cohorts. Further investigations were implemented to determine the associations of the model and inflammatory response–related genes with immune cell infiltration and the responses to chemotherapeutic, targeted, and immunotherapeutic agents. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-128/rc).
Methods
Study design
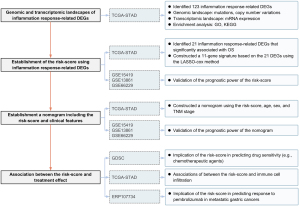
First, the mRNA expression levels of 1,153 inflammatory response–related genes were compared between the tumoral and normal samples from TCGA-STAD cohort. The differentially expressed genes (DEGs) were identified for the construction of the prognostic model. Second, a prognostic signature was developed in the TCGA-STAD cohort and validated in 3 independent cohorts (GSE15459, GSE13861, and GSE66229); its prognostic effect was further explored in the TCGA pan-cancer data sets. Third, a nomogram involving the risk score, age, sex, and tumor stage was constructed in the TCGA-STAD cohort and validated in 3 cohorts (GSE15459, GSE13861, and GSE66229). Fourth, the association of the risk score with drug sensitivity, TME, clinical characteristics, and gene mutation was analyzed in TCGA-STAD cohort. Finally, the ability of the risk score to predicting immunotherapy efficacy was examined in the ERP107734 cohort. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The flowchart of this study is shown in Figure 1.
Data collection
Transcriptomic and clinical data of patients with GC were downloaded from the University of California Santa Cruz (UCSC) Xena (TCGA-STAD, 348 tumor and 31 nontumor tissues), the Gene Expression Omnibus (GEO) database (GSE15459, 190 tumors; GSE13861, 64 tumors; GSE66229, 300 tumors), and the European Nucleotide Archive (ENA) database (ERP107734, 45 patients with advanced GC receiving pembrolizumab monotherapy). Patients with gene expression matrix and prognostic information were included in this study. Overall survival (OS) was reported as the prognostic outcomes in the training and validation datasets. Clinical features including age, sex, tumor stage, EBV infection status, microsatellite instability (MSI) status, TP53 mutation, H. pylori infection status, radiation therapy, and race were described in the TCGA-STAD cohort, while the other 3 validation cohorts provided only the age, sex, and tumor stage information. The baseline characteristics of the samples in the training and validation data sets are described in Table S1. Immunohistochemistry (IHC) staining samples were obtained from The Human Protein Atlas (HPA) database (Human Protein Atlas v22.0. proteinatlas.org, https://www.proteinatlas.org/) (41). All used data are publicly available and were analyzed following the data access policies and publication guidelines. The 1,153 inflammatory response–related genes analyzed in our study were retrieved from the National Center of Biotechnology Information (NCBI) gene database with the keyword being “inflammatory response” and the filter being “Homo sapiens.” The genomic indices including TMB, neoantigens, number of segments, fraction altered, and aneuploidy score were retrieved from a pan-cancer TCGA study (42).
Construction of the prognostic signature based on inflammatory response–related genes
In TCGA cohort, the “limma” package in R (The R Foundation of Statistical Computing) was used to identify the DEGs between tumor tissues and nontumor tissues with a threshold of |log2(fold change)|≥1 and an adjusted P value (Q value) <0.05 (43). A univariable Cox analysis of overall survival (OS) was performed to screen inflammatory response–related genes with a prognostic value. The LASSO-penalized Cox regression analysis was applied to construct a prognostic model with the “glmnet” package in R (44). The risk scores of patients were calculated according to the log2 [fragments per kilobase million (FPKM) + 1] of each gene and its corresponding regression coefficients as follows:
Here, n is the number of prognostic genes, expi is the expression level of gene i, and βi refers to the regression coefficient of gene i. The risk score was constructed in the training cohort (TCGA-STAD cohort) and calculated with the fixed equation for validation in the GSE15459, GSE13861, and GSE66229 cohorts.
Univariable and multivariable analyses were implemented to evaluate the independence of the risk score and identify clinical features for nomogram development. The common clinical factors identified in the univariable analysis across the training and validation sets were involved in the construction and validation of the nomogram. The nomogram was constructed using the R package “survival” in TCGA cohort and validated in the GSE15459, GSE13861, and GSE66229 data sets.
Enrichment analysis
The R package “clusterProfiler” (45) was used to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses based on the inflammatory response–related DEGs [|log2 fold change|≥1 and false discovery rate (FDR) <0.05].
Estimation of immune cell infiltration in the TME
To interpret the composition of the tumor microenvironment, we used cell-type identification by estimating relative subsets of RNA transcript x (CIBERSORTx) to calculate the fraction of 22 types of immune cells in the tumor tissues (46). CIBERSORTx is considered to have better compare to previous deconvolution methods for the analysis of complicated mixtures.
Drug sensitivity analysis
Based on the genomics of drug sensitivity in cancer (GDSC) database, we used the “pRRophetic” package in R to evaluate the susceptibility of patients with GC to chemotherapeutic and targeted agents (47).
Statistical analysis
To assess the between-group differences, we used (I) the Wilcoxon test for continuous variables and (II) Kaplan-Meier curves, log-rank test, and Cox proportional-hazards regression model [hazard ratio (HR) and the 95% CI] for the time-to-event variables. The variables with a P value below 0.10 in the univariable analysis were included in the following multivariable model. The time-dependent area under the receiver operator characteristic (ROC) curve (AUC) was used to assess the prognostic performance and to compare the performances of the current model and previously published models. Spearman correlation was used to test the correlations between continuous variables. Multiple corrections of P values were performed using the Benjamini-Hochberg method. All statistical analyses were performed with R software (version 4.0.0) or SPSS (version 23.0, IBM Corp.). A two-sided P value less than 0.05 was considered statistically significant unless otherwise specified. A predictive model with an AUC value >0.70 was considered as a good predictive efficacy.
Results
Identification of prognostic inflammatory response–related DEGs in TCGA cohort
Of the 1,153 inflammatory response–related genes identified from the NCBI database, 123 were expressed differently between the tumor and the nontumor tissues of TCGA cohort (Figure 2A). The chromosomal loci of these genes are shown in Figure 2B. Somatic mutations of these inflammatory response–related genes are shown in Figure 2C. TP53 mutations were observed in 46% of patients, and other genes were mutated in a small fraction of the patients.
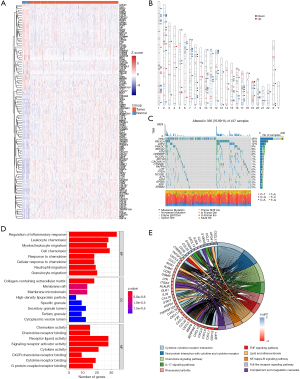
To elucidate the biological functions of the inflammatory response–related DEGs, GO enrichment and KEGG pathway analyses were performed. As expected, these DEGs were enriched in inflammation-related biological processes, especially the chemotaxis and migration of myeloid leukocytes (Figure 2D). Being enriched in membrane organelles, these genes may involve the binding of cytokines or chemokines with receptors in the signaling pathways associated with cancer or innate immunity, such as tumor necrosis factor (TNF) signaling, toll-like receptor signaling, and complement cascades (Figure 2E).
Construction and validation of the prognostic model based on the inflammatory response–related genes
Univariable Cox analysis revealed that 21 inflammatory response–related DEGs (treated as continuous variables) were correlated with the OS of TCGA cohort (Figure 3A). When TCGA patients were divided into 2 groups based on the median of the mRNA level of individual genes, significant differences were seen between the survival of the 2 groups (Figure 3B,3C). LASSO Cox regression analysis was applied to analyze the association of OS with the expression of these 21 genes, and an 11-gene prognostic model based on the optimal λ valve was established (Figure 3D,3E). The 11 genes that most contributed to the prognostic model were SERPINE1, CREB3L3, ADAMTS12, APOD, GFRA1, KIT, ZFP36, APOA1, DNMT1, PVT1, and TNFAIP2, respectively (Table S2). The representative images concerning the IHC staining of the proteins coded by these genes are downloaded from the Human Protein Atlas database (Human Protein Atlas v22.0. proteinatlas.org, https://www.proteinatlas.org/) and shown in Figure S1. ADAMTS12, APOD, and GFRA1 were enriched in fibroblasts, while APOA1 and CREB3L3 were enriched in tumor cells. SERPINE1 was observed in both fibroblasts and tumor cells, while DNMT, TNFAIP2, and ZFP36 were observed in fibroblasts, tumor cells, and tumor-infiltrating immune cells. In contrast, the protein of KIT was not detected in any cell types.
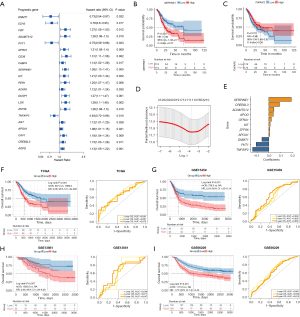
According to the median of risk score, TCGA patients were stratified into a high-risk group (n=174) or a low-risk group (n=174). The high-risk group had a significantly worse OS than did the low-risk group (HR =2.08, 95% CI: 1.49–2.90; P<0.001; Figure 3F). The performance of the risk score was further evaluated by time-dependent ROC curves, and the 1-, 3-, and 5-year AUCs were 0.691, 0.667, and 0.678, respectively (Figure 3F).
To test the robustness of this model, 3 independent data sets including the GSE15459 (n=190), GSE13861 (n=64), and GSE66229 (n=300) were used as the validation cohorts. The risk score of each individual was calculated with the regression coefficient values of the 11 genes (Table S2) according to the equation mentioned above. The high-risk and low-risk groups were defined according to the median value in each cohort. The high-risk patients also had a shorter OS compared with their low-risk counterparts (GSE15459: HR =2.04, 95% CI: 1.33–3.14, P<0.001; GSE13861: HR =2.85, 95% CI: 1.29–6.28, P=0.007; GSE66229: HR =1.75, 95% CI: 1.27–2.42, P<0.001; Figure 3G-3I). The 1-, 3-, and 5-year AUCs were 0.602, 0.602, and 0.650 in the GSE15459 cohort, 0.648, 0.611, and 0.647 in the GSE13861 cohort, and 0.661, 0.630, and 0.610 in the GSE66229 cohort, respectively (Figure 3G-3I).
Univariable and multivariable Cox regression analyses were implemented in TCGA and GEO cohorts (Table 1). After adjustment for the confounding factors, the association between the risk score and OS remained significant in the multivariable models (TCGA: multivariable HR =2.35, 95% CI: 1.49–3.71, P<0.001; GSE15459: multivariable HR =2.10, 95% CI: 1.37–3.24, P=0.001; GSE13861: multivariable HR =2.54, 95% CI: 1.15–5.60, P=0.02; GSE66229: multivariable HR =1.55, 95% CI: 1.12–2.14, P=0.008).
Table 1
| Parameter | Univariable analysis | Multivariable analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| TCGA gastric cancer cohort | |||||
| Age (≥65 vs. <65 years) | 1.54 (1.09–2.17) | 0.01 | 1.34 (0.86–2.10) | 0.19 | |
| Sex (male vs. female) | 1.35 (0.95–1.94) | 0.10 | 1.16 (0.74–1.82) | 0.53 | |
| Tumor stage (III/IV vs. I/II) | 1.81 (1.27–2.58) | 0.001 | 1.56 (1.00–2.43) | 0.05 | |
| EBV infection (positive vs. negative) | 0.94 (0.48–1.85) | 0.86 | |||
| MSI (MSI-H vs. MSI-L/MSS) | 0.73 (0.46–1.16) | 0.19 | |||
| TP53 mutation (mutated vs. wild type) | 0.80 (0.54–1.18) | 0.26 | |||
| H. pylori infection (positive vs. negative) | 0.65 (0.28–1.50) | 0.31 | |||
| Radiation therapy (with vs. without) | 0.32 (0.15–0.66) | 0.002 | 0.36 (0.17–0.75) | 0.007 | |
| Asian (yes vs. no) | 0.70 (0.43–1.15) | 0.16 | |||
| Risk score (high-risk vs. low-risk) | 2.08 (1.49–2.90) | <0.001 | 2.35 (1.49–3.71) | <0.001 | |
| GSE15459 | |||||
| Age (≥65 vs. <65 years) | 0.94 (0.63–1.40) | 0.75 | |||
| Sex (male vs. female) | 1.40 (0.91–2.17) | 0.13 | |||
| Tumor stage (III/IV vs. I/II) | 6.51 (3.60–11.81) | <0.001 | 6.66 (3.67–12.10) | <0.001 | |
| Risk score (high-risk vs. low-risk) | 2.04 (1.33–3.14) | 0.001 | 2.10 (1.37–3.24) | 0.001 | |
| GSE13861 | |||||
| Age (≥65 vs. <65 years) | 1.20 (0.58–2.52) | 0.62 | |||
| Sex (male vs. female) | 1.27 (0.59–2.73) | 0.55 | |||
| Tumor stage (III/IV vs. I/II) | 7.70 (2.32–25.54) | 0.001 | 7.17 (2.15–23.85) | 0.001 | |
| Risk score (high-risk vs. low-risk) | 2.85 (1.30–6.28) | 0.009 | 2.54 (1.15–5.60) | 0.021 | |
| GSE66229 | |||||
| Age (≥65 vs. <65 years) | 1.32 (0.96–1.81) | 0.09 | |||
| Sex (male vs. female) | 1.11 (0.79–1.55) | 0.56 | |||
| Tumor stage (III/IV vs. I/II) | 3.41 (2.34–4.96) | <0.001 | 3.24 (2.22–4.73) | <0.001 | |
| Risk score (high-risk vs. low-risk) | 1.75 (1.27–2.42) | 0.001 | 1.55 (1.12–2.14) | 0.008 | |
CI, confidence interval; HR, hazard ratio; TCGA, The Cancer Genome Atlas; EBV, Epstein-Barr virus; MSI, microsatellite instability; MSI-H, microsatellite instability-high; MSI-L, microsatellite instability-low; MSS, microsatellite stability. H. pylori, Helicobacter pylori.
Compared with other previously published models (12,14,15), the risk score exhibited a higher concordance index (proposed risk score: 0.654; Qing et al.: 0.584; Wang et al.: 0.572; Song et al.: 0.576) and numerically better AUCs in predicting the 1-, 3-, and 5-year OS (Figure S2A-S2C). The exploratory analysis of the prognostic power of the risk score in other cancer types was implemented using TCGA data. A high risk score was associated with shorter OS in patients with mesothelioma (HR =2.38, 95% CI: 1.44–3.92; P<0.001), brain lower grade glioma (HR =1.88, 95% CI: 1.28–2.76; P<0.001), bladder urothelial carcinoma (HR =1.71, 95% CI: 1.26–2.30; P<0.001), thyroid carcinoma (HR =3.91, 95% CI: 1.11–13.75; P=0.022), lung squamous cell carcinoma (HR =1.45, 95% CI: 1.10–1.90; P=0.008), sarcoma (HR =1.56, 95% CI: 1.04–2.32; P=0.029), or breast invasive carcinoma (HR =1.39, 95% CI: 1.00–1.93; P=0.048; Figure S3). In comparison, a high-risk score was linked with a longer OS in thymoma (HR =0.10, 95% CI: 0.01–0.83; P=0.010; Figure S3).
Construction and validation of the nomogram based on the risk score and key clinical features
According to the results of univariable analysis, clinical features including age, sex, and tumor stage were involved in the construction of a nomogram, together with the risk score (Figure 4A). Together with the risk score (the AUC for 1-, 3-, 5-year in TCGA-STAD cohort: 0.691, 0.644, and 0.707; GSE15459: 0.602, 0.602, and 0.650; GSE13861: 0.648, 0.611, and 0.647; GSE66229: 0.661, 0.630, and 0.610), clinical features including age (TCGA-STAD cohort: 0.585, 0.585, and 0.555; GSE15459: 0.516, 0.460, and 0.454; GSE13861: 0.505, 0.595, and 0.547; GSE66229: 0.550, 0.520, and 0.525), sex (TCGA-STAD cohort: 0.522, 0.469, and 0.525; GSE15459: 0.503, 0.592, and 0.594; GSE13861: 0.553, 0.502, and 0.436; GSE66229: 0.445, 0.473, and 0.494), and tumor stage (TCGA-STAD cohort: 0.571, 0.549, and 0.562; GSE15459: 0.697, 0.782, and 0.798; GSE13861: 0.695, 0.745, and 0.747; GSE66229: 0.680, 0.691, and 0.668) were involved in the nomogram. The 1-, 3-, and 5-year AUCs of the nomogram were 0.759, 0.706, and 0.742 in TCGA-STAD cohort, respectively (Figure 4B), with the calibration curves showing a robust prediction of 1-, 3-, and 5-year OS (Figure 4B). The nomogram was validated in the GSE15459 (1-, 3-, and 5-year AUCs were 0.773, 0.786, and 0.803, respectively; Figure 4C), the GSE13861 (1-, 3-, and 5-year AUCs were 0.749, 0.881, and 0.795, respectively; Figure 4D), and the GSE66229 (1-, 3-, and 5-year AUCs were 0.773, 0.735, and 0.722, respectively; Figure 4E). Compared with single clinical signatures and the risk score, the nomogram exhibited improved predictive accuracy in both the training and validation datasets (Table 2). These results suggest that the combination of the risk score and clinical features may be a robust tool for risk stratification in patients with GC.
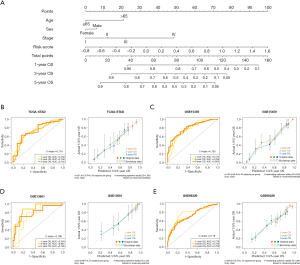
Table 2
| Prognostic signatures | 1-year OS AUC | 3-year OS AUC | 5-year OS AUC |
|---|---|---|---|
| TCGA gastric cancer cohort | |||
| Sex | 0.522 | 0.469 | 0.525 |
| Age | 0.585 | 0.585 | 0.555 |
| Tumor stage | 0.571 | 0.549 | 0.562 |
| Risk score | 0.691 | 0.644 | 0.707 |
| Nomogram | 0.759 | 0.706 | 0.742 |
| GSE15459 | |||
| Sex | 0.503 | 0.592 | 0.594 |
| Age | 0.516 | 0.460 | 0.454 |
| Tumor stage | 0.697 | 0.782 | 0.798 |
| Risk score | 0.602 | 0.602 | 0.650 |
| Nomogram | 0.773 | 0.786 | 0.803 |
| GSE13861 | |||
| Sex | 0.553 | 0.502 | 0.436 |
| Age | 0.505 | 0.595 | 0.547 |
| Tumor stage | 0.695 | 0.745 | 0.747 |
| Risk score | 0.648 | 0.611 | 0.647 |
| Nomogram | 0.749 | 0.881 | 0.795 |
| GSE66229 | |||
| Sex | 0.445 | 0.473 | 0.494 |
| Age | 0.550 | 0.520 | 0.525 |
| Tumor stage | 0.680 | 0.691 | 0.668 |
| Risk score | 0.661 | 0.630 | 0.610 |
| Nomogram | 0.773 | 0.735 | 0.722 |
TCGA, The Cancer Genome Atlas; OS, overall survival; AUC, area under curve.
Correlation of the risk score with the clinical characteristics of patients
In the analysis of the association between the risk score and clinical characteristics of patients with GC, no significant difference was observed between patients with different sex, age, tumor stage, or of Helicobacter pylori infection status (Figure S4A-S4D), while the patients who did not receive radiation therapy had a significantly higher risk score than those who did (Figure S4E).
The risk score also varied among molecular types. The genome stable subtype had a higher risk score than did the chromosomal instability (CIN), Epstein-Barr virus (EBV)–positive, and MSI subtypes, while the MSI subtype showed the lowest median of the risk score (Figure S4F). The low-risk group and the high-risk group held similar somatic mutation patterns, but the former tended to have a higher mutation frequency (Figure S4G-S4H) and also significantly higher silent and nonsilent mutation rates and higher single-nucleotide variant neoantigens (Figure S4I).
Associations of the inflammatory response–related genes with treatment responses
First, we analyzed the correlations of the risk score with the sensitivities to common antitumor drugs (Figure 5A and Table S3). For cisplatin and docetaxel, the risk score showed no significant correlation with the half maximal inhibitory concentration (IC50) values. However, the sensitivity of tumor cells to some other chemotherapy drugs, including mitomycin-C and vinorelbine, slightly decreased with the increase of the risk score. A negative correlation was revealed between the risk score and the IC50 values of the drugs targeting the BCR–ABL fusion protein, sarcoma (Src), vascular endothelial growth factor (VEGFR), and phosphatidylinositol-3-kinase (PI3K)–mammalian target of rapamycin (mTOR) pathway. However, the IC50 values of several inhibitors of epidermal growth factor receptor (EGFR) or polo-like kinase (PLK) were positively correlated with the risk score, especially afatinib (BIBW2992).
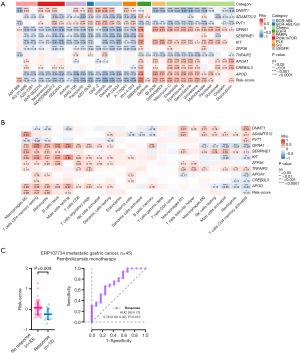
Considering that the activated TME may lead to better survival outcomes, especially for patients treated with immune checkpoint inhibitors (ICIs), we quantified the fractions of 22 immune cell subpopulations in the tumor samples (Table S4). In the low-risk group, there were more activated CD4+ memory T cells and follicular helper T cells, while more naïve B cells, monocytes, and resting mast cells were observed in the high-risk group (Figure S5). Correlation analysis also revealed that the risk score was positively correlated with the densities of monocytes, M2 macrophages, resting CD4+ memory T cells, and resting mast cells, while it was negatively correlated with activated CD4+ memory T cells (Figure 5B). The expressions of most of the candidate DEGs were significantly related to the infiltration of activated/resting CD4+ memory T cells, M2 macrophage, and monocytes. These results suggest that the tumors with a higher risk score may have an immunosuppressive microenvironment, which may impede the immune surveillance and elimination of the cancer cells.
Subsequently, the ERP107734 cohort including 45 patients with advanced GC treated with pembrolizumab monotherapy was used to estimate the association between the risk score and response to ICIs. Compared to the nonresponders (n=33), the responders (n=12) had a significantly lower risk score (P=0.009; Figure 5C). We plotted the ROC curve for predicting the patient’s response to pembrolizumab based on the risk score and found an AUC of 0.755 (95% CI: 0.595–0.915; P=0.010; Figure 5C).
Discussion
Inflammation, which enables epigenetic alterations and results in the production of growth factors, is a crucial cause of newly emergent tumors and malignant progression. Inflammation-reducing strategies that inhibit either the initiation or propagation of persistent inflammation (e.g., anti-infective agents, nonsteroidal anti-inflammatory drugs (NSAIDs), and other inflammation-reducing drugs including statins and metformin) might therefore prevent or delay cancer initiation (48). In gastric cancer, antiviral therapies for EBV and antibacterial therapies for H. pylori are the main anti-inflammatory strategies in GC treatment. Besides, targeting interleukin-6 may overcome stroma-induced resistance to chemotherapy in GC (49). However, some pro-inflammatory cytokines or stimulators (TNF-α, cGAS-STING pathway activators) can promote the infiltration of immune cells into infected tissues and thus significantly improve the efficacy of tumor therapy (50), suggesting that inflammation is a “double-edged sword”, making inflammation regulation an important issue to improve the efficacy of cancer therapy (51). Identification of the most critical drivers affecting inflammatory TME tumor, avoidance of the conversion of acute-to-chronic inflammation induced by anticancer therapies, and reduction of the severe inflammatory side effects of anticancer therapies (CAR-T therapy induced inflammatory storm) are important challenges involved in inflammation regulation (51,52). In addition, the different inflammatory responses of cancer patients should be considered in cancer management, and personalized treatment strategies regarding tumor-associated inflammation will help improve anti-cancer efficacy.
In this study, we systematically investigated the mRNA expression of inflammatory response–related genes in GC tissues and analyzed their associations with OS. A novel prognostic model integrating 11 inflammatory response–related genes was constructed based on TCGA GC data and validated in 3 independent cohorts. Further analyses revealed that the risk score obtained by our model may represent the pattern of the TME constructed through the recruitment and differentiation of various immune cells. A high risk score indicated the presence of an immunosuppressive microenvironment, potentially leading to patients’ nonresponse to immunotherapy, which is supported by the results of ERP107734 cohort. Moreover, the risk score was related to the tumor cell’s sensitivity to chemotherapeutic and targeted drugs.
The relationship between inflammation and cancer has been well studied, and cancer prognosis estimation based on inflammatory response–related factors, such as with the Glasgow prognostic score and its modified version, has been conducted (31). Although the early studies in this area were limited to a few typical factors, such as C-reactive protein, albumin, and lymphocyte–monocyte ratio, the development of high-throughput sequencing technology in the last decade has made it possible to comprehensively screen hub biomarkers. We first analyzed the correlation between the expression of multiple inflammatory response–related genes and the survival outcomes of patients with GC. A weighted model to estimate the prognosis was established, and its independence and robustness were then verified in a training cohort and an external validation cohort. Several candidate genes in our model, including ADAMTS12, APOA1, APOD, and ZFP36, have been reported to be inflammation inhibitors (53-57) and were associated with a high-risk score in our study. On the other hand, PVT1, DNMT1, and TNFAIP2 have been reported to be proinflammatory genes (58-64), and their expressions were negatively correlated with the risk score in our study. Overall, our model attested to the role of inflammatory response in cancer prognosis, highlighting the importance of inflammatory response in the suppression of the tumor.
In addition to the association between GC prognosis and mRNA levels of inflammatory-related genes, previous study has also explored the role of gene polymorphisms in GC. For instance, the rs2227692 C>T polymorphism in SERPINE1 intron affecting gene expression is associated with diffuse-type gastric cancer susceptibility (65). The TNFAIP2 3' UTR rs8126 T>C polymorphism, which might affect TNFAIP2 protein expression, is associated with GC risk in the Chinese population, especially in cases with males aged 60 years or older, H. pylori-negative, non-smoking and non-drinking individuals (66). Besides, several researches and meta-analyses have also demonstrated the association between the DNMT1 rs2228612 A>G polymorphism and GC risk (67-69). These results emphasized the potential roles of inflammatory-related gene polymorphisms in GC, and the associations of polymorphisms of CREB3L3, ADAMTS12, APOD, GFRA1, KIT, ZFP36, APOA1, and PVT1 with GC risk are of great potential for further researches.
Although the risk score was trained in TCGA-STAD cohort with a majority Western population, it was validated in both the Asian population cohorts (GSE15459 and GSE66229) and the Western population cohort (GSE13861), indicating the race-independent prognostic power of the risk score. The combination of the risk score and clinical features including age, sex, and stage achieved consistent prognostic power across all cohorts, suggesting that the risk score may be a robust and useful tool for risk stratification in patients with GC.
Various immune cells involved in inflammatory response constitute the complicated immune microenvironment, which may affect the immune surveillance and elimination of tumor cells during treatment, especially immunotherapy. In the present study, patients with a high-risk score tended to have a poor survival outcome, an immunosuppressive TME, and an unfavorable response to pembrolizumab monotherapy. These results indicate the potential of predicting immunotherapy efficacy using the activity of inflammatory response in advanced GC. We also found that the tumor cells with a high-risk score may be resistant to several chemotherapeutic drugs and sensitive to the chemicals targeting BCR–ABL, Src, PI3K/mTOR, and VEGFR. These results suggest the possibility of applying this model in more scenarios.
TCGA project has uncovered four molecular subtypes of gastric cancer: EBV, MSI, genomically stable (GS), and CIN. Based on our results, the EBV and MSI subtypes obtained lower risk scores compared to the CIN and GS subtypes (Figure S4F), which was associated with improved prognosis. These results coincide with previously published studies reporting the worst prognosis in the GS subtype and the best prognosis in the EBV subtype (70). Besides, the lower risk scores in the EBV and MSI subtypes were associated with lower densities of immunosuppressive tumor-infiltrating immune cells, lower levels of TGF-beta response, and response to pembrolizumab, suggesting the potential of immunotherapies in EBV and MSI subtypes of GC with lower risk scores. Taken together, hopefully, these results can explain preferably the association between the risk score and the GC molecular subtypes, which may provide evidence for further research into the inflammatory features in GC molecular subtypes. Constructed and validated in 4 independent cohorts, our model exhibited wide utility in different populations. Besides, the measurement of mRNA levels of 11 genes was relatively economic, and together with the easily accessible clinical information, this model is of great potential for clinical practice. However, some limitations to this study should also be mentioned. First, since our model was constructed and validated with limited retrospective public data, more prospective studies are warranted to verify its clinical utility. Second, the links between the risk score and immune activity or drug sensitivity based on in silico prediction need to be experimentally addressed. Third, considering the highly complex TME of GC and multiple driving factors of the GC prognosis, a single model integrating only mRNA data might not be sufficient to guide the clinical decision-making. Although we have developed a nomogram including the risk score and clinical features including age, sex, and stage, the prediction accuracy of this nomogram can be improved if other genomic features are also incorporated. However, the genomic data are only available in TCGA training cohort and are missing in the 3 validation sets. A GC cohort with multiomics data should be included in future research.
Conclusions
Our study constructed a novel model based on the mRNA expression of 11 inflammatory response–related genes in GC. The risk score obtained by this model was proven to be independently associated with survival outcomes in both the training and the validation cohorts, demonstrating the robustness of its prognostic utility. Additionally, the risk score was associated with immune cell infiltration and responses to chemotherapeutic, targeted, and immunotherapeutic treatments. With further validation using a prospective setting, this inflammatory response–related model may potentially change the management of GC by enabling risk stratification and predicting response to systemic therapy.
Acknowledgments
We thank Dizai Shi (Stitch) for his emotional support as well as the included patients and their family members for their understanding and participation.
Funding: This work was supported by 2018 Entrepreneurial Leading Talent of Guangzhou Huangpu District and Guangzhou Development District (No. 2022-L023).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-128/rc
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-128/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-128/coif). TZ, YX, WC, QZ, GW, LL, YH, and SC are employed by Burning Rock Biotech. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635-48. [Crossref] [PubMed]
- Chia NY, Tan P. Molecular classification of gastric cancer. Ann Oncol 2016;27:763-9. [Crossref] [PubMed]
- Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol 2020;18:534-42. [Crossref] [PubMed]
- Yu JI, Lim DH, Lee J, et al. Comparison of the 7th and the 8th AJCC Staging System for Non-metastatic D2-Resected Lymph Node-Positive Gastric Cancer Treated with Different Adjuvant Protocols. Cancer Res Treat 2019;51:876-85. [Crossref] [PubMed]
- Kim SG, Seo HS, Lee HH, et al. Comparison of the Differences in Survival Rates between the 7th and 8th Editions of the AJCC TNM Staging System for Gastric Adenocarcinoma: a Single-Institution Study of 5,507 Patients in Korea. J Gastric Cancer 2017;17:212-9.
- Adachi Y, Yasuda K, Inomata M, et al. Pathology and prognosis of gastric carcinoma: well versus poorly differentiated type. Cancer 2000;89:1418-24. [Crossref] [PubMed]
- Jiang L, Liao J, Han Y. Study on the role and pharmacology of cuproptosis in gastric cancer. Front Oncol 2023;13:1145446. [Crossref] [PubMed]
- Zeng Y, Cai F, Wang P, et al. Development and validation of prognostic model based on extra-gastric lymph nodes metastasis and lymph node ratio in node-positive gastric cancer: a retrospective cohort study based on a multicenter database. Int J Surg 2023; Epub ahead of print. [Crossref] [PubMed]
- Li J, Cui T, Huang Z, et al. Analysis of risk factors for lymph node metastasis and prognosis study in patients with early gastric cancer: A SEER data-based study. Front Oncol 2023;13:1062142. [Crossref] [PubMed]
- Verma R, Sharma PC. Next generation sequencing-based emerging trends in molecular biology of gastric cancer. Am J Cancer Res 2018;8:207-25. [PubMed]
- Xiao W, Ren L, Chen Z, et al. Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat Biotechnol 2021;39:1141-50. [Crossref] [PubMed]
- Cao J, Yang X, Chen S, et al. The predictive efficacy of tumor mutation burden in immunotherapy across multiple cancer types: A meta-analysis and bioinformatics analysis. Transl Oncol 2022;20:101375. [Crossref] [PubMed]
- Xiao C, Dong T, Yang L, et al. Identification of Novel Immune Ferropotosis-Related Genes Associated With Clinical and Prognostic Features in Gastric Cancer. Front Oncol 2022;12:904304. [Crossref] [PubMed]
- Gao W, Yang M. Identification by Bioinformatics Analysis of Potential Key Genes Related to the Progression and Prognosis of Gastric Cancer. Front Oncol 2022;12:881015. [Crossref] [PubMed]
- Wang Z, Cao L, Zhou S, et al. Construction and Validation of a Novel Pyroptosis-Related Four-lncRNA Prognostic Signature Related to Gastric Cancer and Immune Infiltration. Front Immunol 2022;13:854785. [Crossref] [PubMed]
- Yin J, Che G, Wang W, et al. Investigating the Prognostic Significance of Pyroptosis-Related Genes in Gastric Cancer and Their Impact on Cells' Biological Functions. Front Oncol 2022;12:861284. [Crossref] [PubMed]
- Song J, Wei R, Huo S, et al. Metastasis Related Epithelial-Mesenchymal Transition Signature Predicts Prognosis and Response to Immunotherapy in Gastric Cancer. Front Immunol 2022;13:920512. [Crossref] [PubMed]
- Qing X, Xu W, Liu S, et al. Molecular Characteristics, Clinical Significance, and Cancer Immune Interactions of Angiogenesis-Associated Genes in Gastric Cancer. Front Immunol 2022;13:843077. [Crossref] [PubMed]
- Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019;51:27-41. [Crossref] [PubMed]
- Chen D, Lai J, Cheng J, et al. Predicting peritoneal recurrence in gastric cancer with serosal invasion using a pathomics nomogram. iScience 2023;26:106246. [Crossref] [PubMed]
- Nieder C, Mehta MP, Geinitz H, et al. Prognostic and predictive factors in patients with brain metastases from solid tumors: A review of published nomograms. Crit Rev Oncol Hematol 2018;126:13-8. [Crossref] [PubMed]
- Shalapour S, Karin M. Pas de Deux: Control of Anti-tumor Immunity by Cancer-Associated Inflammation. Immunity 2019;51:15-26. [Crossref] [PubMed]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010;140:883-99. [Crossref] [PubMed]
- Lin JX, Lin JP, Xie JW, et al. Prognostic Value and Association of Sarcopenia and Systemic Inflammation for Patients with Gastric Cancer Following Radical Gastrectomy. Oncologist 2019;24:e1091-101. [Crossref] [PubMed]
- Kudou K, Nakashima Y, Haruta Y, et al. Comparison of Inflammation-Based Prognostic Scores Associated with the Prognostic Impact of Adenocarcinoma of Esophagogastric Junction and Upper Gastric Cancer. Ann Surg Oncol 2021;28:2059-67. [Crossref] [PubMed]
- Aurello P, Tierno SM, Berardi G, et al. Value of preoperative inflammation-based prognostic scores in predicting overall survival and disease-free survival in patients with gastric cancer. Ann Surg Oncol 2014;21:1998-2004. [Crossref] [PubMed]
- Li S, Lan X, Gao H, et al. Systemic Inflammation Response Index (SIRI), cancer stem cells and survival of localised gastric adenocarcinoma after curative resection. J Cancer Res Clin Oncol 2017;143:2455-68. [Crossref] [PubMed]
- Liu X, Sun X, Liu J, et al. Preoperative C-Reactive Protein/Albumin Ratio Predicts Prognosis of Patients after Curative Resection for Gastric Cancer. Transl Oncol 2015;8:339-45. [Crossref] [PubMed]
- Pan QX, Su ZJ, Zhang JH, et al. A comparison of the prognostic value of preoperative inflammation-based scores and TNM stage in patients with gastric cancer. Onco Targets Ther 2015;8:1375-85. [Crossref] [PubMed]
- McMillan DC. The systemic inflammation-based Glasgow Prognostic Score: a decade of experience in patients with cancer. Cancer Treat Rev 2013;39:534-40. [Crossref] [PubMed]
- Fu S, Yan J, Tan Y, et al. Prognostic value of systemic immune-inflammatory index in survival outcome in gastric cancer: a meta-analysis. J Gastrointest Oncol 2021;12:344-54. [Crossref] [PubMed]
- Zhao H, Wu L, Yan G, et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct Target Ther 2021;6:263. [Crossref] [PubMed]
- Kroemer G, Galluzzi L, Kepp O, et al. Immunogenic cell death in cancer therapy. Annu Rev Immunol 2013;31:51-72. [Crossref] [PubMed]
- Krysko DV, Garg AD, Kaczmarek A, et al. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer 2012;12:860-75. [Crossref] [PubMed]
- Barker HE, Paget JT, Khan AA, et al. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer 2015;15:409-25. [Crossref] [PubMed]
- Karagiannis GS, Condeelis JS, Oktay MH. Chemotherapy-Induced Metastasis: Molecular Mechanisms, Clinical Manifestations, Therapeutic Interventions. Cancer Res 2019;79:4567-76. [Crossref] [PubMed]
- Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol 2015;12:584-96. [Crossref] [PubMed]
- Zitvogel L, Galluzzi L, Kepp O, et al. Type I interferons in anticancer immunity. Nat Rev Immunol 2015;15:405-14. [Crossref] [PubMed]
- Spiotto M, Fu YX, Weichselbaum RR. The intersection of radiotherapy and immunotherapy: mechanisms and clinical implications. Sci Immunol 2016;1:EAAG1266. [Crossref] [PubMed]
- Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2018;48:812-830.e14. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010;33:1-22. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773-82. [Crossref] [PubMed]
- Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014;9:e107468. [Crossref] [PubMed]
- Hou J, Karin M, Sun B. Targeting cancer-promoting inflammation - have anti-inflammatory therapies come of age? Nat Rev Clin Oncol 2021;18:261-79. [Crossref] [PubMed]
- Ham IH, Oh HJ, Jin H, et al. Targeting interleukin-6 as a strategy to overcome stroma-induced resistance to chemotherapy in gastric cancer. Mol Cancer 2019;18:68. [Crossref] [PubMed]
- Liu Y, Fei Y, Wang X, et al. Biomaterial-enabled therapeutic modulation of cGAS-STING signaling for enhancing antitumor immunity. Mol Ther 2023; Epub ahead of print. [Crossref] [PubMed]
- Zhang Z, Li X, Wang Y, et al. Involvement of inflammasomes in tumor microenvironment and tumor therapies. J Hematol Oncol 2023;16:24. [Crossref] [PubMed]
- Davila ML, Riviere I, Wang X, et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med 2014;6:224ra25. [Crossref] [PubMed]
- Wei J, Richbourgh B, Jia T, et al. ADAMTS-12: a multifaced metalloproteinase in arthritis and inflammation. Mediators Inflamm 2014;2014:649718. [Crossref] [PubMed]
- Mohamedi Y, Fontanil T, Cal S, et al. ADAMTS-12: Functions and Challenges for a Complex Metalloprotease. Front Mol Biosci 2021;8:686763. [Crossref] [PubMed]
- Cochran BJ, Ong KL, Manandhar B, et al. APOA1: a Protein with Multiple Therapeutic Functions. Curr Atheroscler Rep 2021;23:11. [Crossref] [PubMed]
- Rassart E, Desmarais F, Najyb O, et al. Apolipoprotein D. Gene 2020;756:144874. [Crossref] [PubMed]
- Makita S, Takatori H, Nakajima H. Post-Transcriptional Regulation of Immune Responses and Inflammatory Diseases by RNA-Binding ZFP36 Family Proteins. Front Immunol 2021;12:711633. [Crossref] [PubMed]
- Fu J, Shi H, Wang B, et al. LncRNA PVT1 links Myc to glycolytic metabolism upon CD4+ T cell activation and Sjögren's syndrome-like autoimmune response. J Autoimmun 2020;107:102358. [Crossref] [PubMed]
- Zeni PF, Mraz M. LncRNAs in adaptive immunity: role in physiological and pathological conditions. RNA Biol 2021;18:619-32. [Crossref] [PubMed]
- Wu L, Xia J, Li D, et al. Mechanisms of M2 Macrophage-Derived Exosomal Long Non-coding RNA PVT1 in Regulating Th17 Cell Response in Experimental Autoimmune Encephalomyelitisa. Front Immunol 2020;11:1934. [Crossref] [PubMed]
- Wang X, Cao Q, Yu L, et al. Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity. JCI Insight 2016;1:e87748. [Crossref] [PubMed]
- Tang RZ, Zhu JJ, Yang FF, et al. DNA methyltransferase 1 and Krüppel-like factor 4 axis regulates macrophage inflammation and atherosclerosis. J Mol Cell Cardiol 2019;128:11-24. [Crossref] [PubMed]
- Yu J, Qiu Y, Yang J, et al. DNMT1-PPARγ pathway in macrophages regulates chronic inflammation and atherosclerosis development in mice. Sci Rep 2016;6:30053. [Crossref] [PubMed]
- Zhao D, Deng SC, Ma Y, et al. miR-221 alleviates the inflammatory response and cell apoptosis of neuronal cell through targeting TNFAIP2 in spinal cord ischemia-reperfusion. Neuroreport 2018;29:655-60. [Crossref] [PubMed]
- Ju H, Lim B, Kim M, et al. SERPINE1 intron polymorphisms affecting gene expression are associated with diffuse-type gastric cancer susceptibility. Cancer 2010;116:4248-55. [Crossref] [PubMed]
- Guo F, Xu Q, Lv Z, et al. Correlation Between TNFAIP2 Gene Polymorphism and Prediction/Prognosis for Gastric Cancer and Its Effect on TNFAIP2 Protein Expression. Front Oncol 2020;10:1127. [Crossref] [PubMed]
- Li H, Li W, Liu S, et al. DNMT1, DNMT3A and DNMT3B Polymorphisms Associated With Gastric Cancer Risk: A Systematic Review and Meta-analysis. EBioMedicine 2016;13:125-31. [Crossref] [PubMed]
- Li H, Liu JW, Sun LP, et al. A Meta-Analysis of the Association between DNMT1 Polymorphisms and Cancer Risk. Biomed Res Int 2017;2017:3971259. [Crossref] [PubMed]
- Tian J, Liu G, Zuo C, et al. Genetic polymorphisms and gastric cancer risk: a comprehensive review synopsis from meta-analysis and genome-wide association studies. Cancer Biol Med 2019;16:361-89. [Crossref] [PubMed]
- Sohn BH, Hwang JE, Jang HJ, et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin Cancer Res 2017;23:4441-9. [Crossref] [PubMed]
(English Language Editor: J. Gray)


