
KMO in the promotion of tumor development and progression in hepatocellular carcinoma
Highlight box
Key findings
• Kynurenine 3-monooxygenase (KMO) plays an important role in the early diagnosis, prognosis, occurrence, and development of liver cancer, and may target microRNA (miR)-3613-5p to function.
What is known and what is new?
• Microarray analysis has indicated that KMO expression is lower in HCC.
• The mechanism of KMO is unclear in regulating the development of HCC.
What is the implication, and what should change now?
• KMO had a significantly lower expression in HCC tissues and cells. Furthermore, KMO as an oncogene could promote HCC cell proliferation, migration, invasion, apoptosis, and epithelial-mesenchymal transition (EMT). These findings indicated that KMO may be candidate gene in HCC development and progression.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers and an important medical problem with poor prognosis. In 2020, the latest global statistics showed that the number of new cases of liver cancer was 905,677, accounting for 4.7% of all new cancer cases, and the number of deaths was 830,180, accounting for 8.3% all cancer deaths. HCC is ranked as the sixth most common neoplasm and the third leading cause of cancer death (1). The development of HCC is closely related to the presence of chronic liver disease, and the risk factors underlying HCC pathogenesis are highly variable, which is mainly related to hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, aflatoxin B1 exposure, or nonalcoholic fatty liver disease (2,3). There are many traditional treatments for HCC, such as curative resection, liver transplantation, radiofrequency ablation, and systemic targeted agents (e.g., sorafenib), but the treatment of advanced HCC has remained a challenge for many years (4,5). Although there have been advances in surgery and other strategies, the prognosis of patients with HCC is still not optimistic due to the typically late diagnosis of HCC or the advanced stage at the time of diagnosis. Therefore, it is urgent to find an early diagnostic method and elucidate the molecular pathogenesis of HCC.
Recently, due to advances in microarray technology based on high-throughput platforms, bioinformatics analysis has been widely used to screen and identify key biomarkers and potential molecular mechanisms for certain cancers, and use of this technology has been shown to improve overall survival (OS) by up to 5 years. The Gene Expression Omnibus (GEO) online database is a public repository available worldwide for gene expression data sets, as well as original series and platform records (6). We downloaded and reanalyzed 2 original microarray data sets, GSE101728 and GSE88839, from the GEO database and found biomarkers and disease mechanisms that may be valuable for future research. Then, we identified kynurenine 3-monooxygenase (KMO) as a potential gene for HCC patients that was significantly underexpressed in HCC tissues and associated with the poor prognosis of patients with HCC.
KMO encodes a mitochondrial outer membrane protein that catalyzes the hydroxylation of a L-tryptophan metabolite, L-kynurenine, to form L-3-hydroxykynurenine. Numerous studies have shown that KMO plays a central role in the tryptophan metabolism, and KMO has been identified as the main pathogenic factor in neurodegenerative diseases (7-9). Recently, KMO was revealed to a known prognostic marker in human HCC, and some lymphomas and endometrial cancers exhibit moderate to strong KMO cytoplasmic immunoreactivity (7,10,11). In a summary, these studies strongly suggest that KMO is associated with the occurrence of HCC. However, little research has been conducted on the molecular mechanisms of KMO on HCC.
In the study, KMO was identified as the hub gene. To more deeply understand the effect of KMO in the progression of HCC, the mechanism of KMO was investigated in HCC cells. The expression of KMO was obviously inhibited in HCC cells and targeted microRNA (miR)-3613-5p. Finally, KMO was also shown to regulate the epithelial-mesenchymal transition (EMT) progression and the development of HCC in cells. Overall, our data provide insights into the diagnosis and treatment of HCC. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-147/rc).
Methods
Microarray data
In this study, data were download from the online GEO database, which is a publicly accessible, functional genomics data repository supporting high-throughput gene expression data, chips, and microarrays (6). We obtained the gene expression profiles of GSE101728 and GSE88839. GSE101728 contains paired HCC tumor and normal samples from 7 patients, from which we analyzed the expression of messenger RNA (mRNA); GSE88839 contained 35 HCC tumor tissue and 3 normal liver tissue, from which the expression of mRNA was determined. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Microarray data analysis
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) is an online web software used for screening differentially expressed genes (DEGs) by comparing samples from GEO series. In our study, GEO2R was used to search DEGs between HCC and normal control samples. |Log fold change (FC)| ≥1 and P<0.05 were defined as the screening criteria for DEGs. Volcano maps were produced using the “ggplot2” package in R software (The R Foundation of Statistical Computing) with the screening criteria |log(FC)| ≥1 and P<0.05. Subsequently, we used Venn online software (http://bioinfogp.cnb.csic.es/tools/venny/index.html) to identify the overlapping DEGs of the 2 data sets.
Function enrichment analysis of DEGs
Gene Ontology (GO) provides an integrated source of digital data relating to the functions of genes (12). GO annotation contains 3 categories: biological process (BP), cellular component (CC), and molecular function (MF). Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database with high-level functional interpretation and the practical application of genomic information (13). The Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.8 (https://david.ncifcrf.gov) (14) is widely used for gene function analysis. In our study, DEGs obtained by Venn online software were enriched by GO function and KEGG pathway with DAVID, and a P value <0.05 was considered statistically significant.
Protein-protein interaction (PPI) network analysis of DEGs
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org) (15) was used to construct the PPI network, which provides critical assessment and integration of protein interactions, including direct and indirect associations. The results of the analysis were visualized in Cytoscape (v3.8.2) software (16). The plug-ins CytoHubba (17) and minimal common oncology data elements (mCODE; degree cutoff =2, κ-score =2, maximum depth =100, node density cutoff =0.1, and node score cutoff =0.2) in Cytoscape were downloaded and installed (18). The top 10 scores or the first cluster was taken as the criterion to screen out the hub genes with high connectivity in the gene expression network.
Analysis of target genes
First, through the Tumor IMmune Estimation Resource (TIMER) (http://timer.cistrome.org/) (19) website, the expression of the target gene in various tumors was found. Furthermore, Gene Expression Profiling Interactive Analysis (GEPIA) (20) (http://gepia.cancer-pku.cn/) and the UALCAN (http://ualcan.path.uab.edu/analysis.html) (21) websites were used to determine the expression of the target gene in HCC.
Identification of microRNA-mRNA pairs
In our study, the microRNA (miRNA) gene pairs of the hub genes were screened using the miRDB (22) (http://mirdb.org/index.html), TarBase v. 8.0 (23) (http://mirtarbase.cuhk.edu.cn/php/index.php), starBase v. 3.0 (24) (http://starbase.sysu.edu.cn/index.php), and TargetScan (http://www.targetscan.org/vert_72/) databases (25); miRNAs appearing in at least 3 of these databases were identified as the potential miRNAs.
Survival analysis of hub mRNAs and miRNAs
A KaplanMeier (K-M) plotter (26) (http://kmplot.com/analysis/index.php?p=background) was used to predict the OS of the genes. To further clarify the relationship between bub mRNA and miRNA expression and HCC prognosis, the K-M plotter was used for survival analysis, while the log-rank test was used for statistical analysis. In the log-rank test, P<0.05 was considered statistically significant. The hub mRNAs and miRNAs were taken as the key genes in HCC prognosis.
Cell lines and culture
The normal liver cell lines HL7702, and liver cancer cell lines MHCC97-L, MHCC97-H, and BEL-7402 were purchased from Beijing BeInnovation Biotechnology Research Institute (BNCC; Beijing, China). The HCC-9903 cell lines were stored in our laboratory. The HL7702 and BEL-7402 cells were cultured in RPMI-1640 with 10% or 20% fetal bovine serum (FBS), while the MHCC97-L, HCC-9903, and MHCC97-H cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS. All cells were cultured in a 37 ℃, 5% CO2 cell incubator.
Plasmid construction and transfection
miR-3613-5p mimics and inhibitors, small interfering KMO (si-KMO), and negative controls were purchased from RiboBio (Guangzhou, China). The transfection was conducted with Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s protocol.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Complementary DNA (cDNA) was reversed with a M-MuLV First Strand cDNA Synthesis Kit (Shanghai Sango Biotechnology Co., Ltd., Shanghai, China). qRT-PCR was carried out using SYBR Green. All primers were synthesized by Shanghai Sango Biotechnology Engineering Co., Ltd. (see the Table 1). The relative expression level of the target gene was calculated using the 2−ΔΔCt method.
Table 1
| Gene name | Primer (forward-reverse) |
|---|---|
| KMO | Forward: 5'-TGTCAACTCAAGCTGGTTCATT-3' Reverse: 5'-TGGCTATCAGTGATCCCAAGAAA-3' |
| miR-3613-5p | Forward: 5'-TGCGGTGTTGTACTTTTTT-3' Reverse: 5'-CCAGTGCAGGGTCC-3' |
PCR, polymerase chain reaction; KMO, kynurenine 3-monooxygenase.
Cell Counting Kit 8 (CCK-8) assay
The cells in the logarithmic lifetime at a density of 2×104 cells/well were vaccinated into a 96-well plate at 100 µL/plate. Cell Counting Kit (CCK) reagent (10 µL) (Beyotime Institute of Biotechnology, Jiangsu, China) was added to each well at 24, 48, 72, and 96 h after incubation, and incubation occurred for another 4 h thereafter. Finally, the plate was removed and placed on the enzyme marker (BioTek Instruments, Winooski, VT, USA) for detection of absorbance, and the absorbance values was recorded at a 450-nm wavelength.
Transwell assay
Transwell assay was applied to analyze cell migration and invasion. Serum-free HCC medium cells at a cell density of 2×106 were added to the upper chamber of the Transwell cell. Matrigel matrix glue was added to the upper chamber of the invasion assay, but not add Matrigel matrix glue in the migration assay. The method was referred as the article (27). Finally, the cells were stained with 0.1% crystal violet for 20 min and observed with an Olympus BX51 microscope (Olympus, Tokyo, Japan) and counted with ImageJ software (US National Institutes of Health, Bethesda, MD, USA).
Western blot assay
First, 10% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) was used to extract and separate cellular proteins, after which the cell proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (MilliporeSigma, Burlington, MA, USA). Second, the membranes were blocked with 5% skim milk in phosphate-buffered saline with Tween20 (PBST) solution and incubated with the primary antibody of the target protein at 4 ℃ overnight. Third, the membrane was incubated with the secondary antibody at room temperature for 1 h and then exposed to an enhanced chemiluminescence (ECL) system (Bio-Rad, Hercules, CA, USA) to detect the protein bands. The antibodies used in this method are listed in Table 2.
Table 2
| Antibody | Supplier | Catalog # | Application |
|---|---|---|---|
| KMO | Abcam | ab167274 | WB (1:1,000) |
| Vimentin | CST | 5741 | WB (1:1,000) |
| N-cadherin | CST | 13116 | WB (1:1,000) |
| E-cadherin | CST | 14472S | WB (1:1,000) |
| Slug | Abcam | ab27568 | WB (1:1,000) |
| Snail | Abcam | ab216347 | WB (1:1,000) |
| β-actin | Abcam | ab8226 | WB (1:1,000) |
| TWIST | Abcam | ab50887 | WB (1:1,000) |
| Goat anti-mouse IgG H&L (HRP) | Abcam | ab205719 | WB (1:5,000) |
| HRP anti-mouse | CST | 7076 | WB (1:5,000) |
| HRP anti-rabbit | CST | 7074 | WB (1:5,000) |
KMO, kynurenine 3-monooxygenase; TWIST, twist family bHLH transcription factor 1; IgG, immunoglobin G; H&L, heavy and light chain; HRP, horseradish peroxidase; CST, Cell Signaling Technology; WB, Western blotting.
Flow cytometry
The liver cancer cells were digested with trypsin without ethylenediaminetetraacetic acid (EDTA), washed twice with cold PBS, and collected by centrifugation. Annexin V-enhanced green fluorescent protein (EGFP) and propidium iodide (5 µL) were added to the cells (1×106 cells/mL), mixed well, and then placed at room temperature without light for 15 minutes. After placement, 400 µL of 1× binding buffer was added, and then flow cytometry was used to detect apoptosis within 1 h.
Statistical analysis
Data were analyzed with SPSS v. 20 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA). Correlations were tested with Spearman rank correlation (Rs). Raw data are expressed as medians with ranges and presented graphically as box plots (showing median and quartiles), with outliers (according to Tukey criteria) indicated separately. Statistical significance was set at P<0.05.
Results
Identification of DEGs in HCC
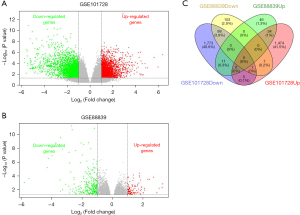
According to the defined criteria, 2 array data sets (GSE101728 and GSE88839) were included in the research. The basic information for the two data sets in the Table 3. A total of 4,335 and 373 DEGs were extracted from GSE101728 and GSE88839 using the GEO2R online tool and visualized with a volcano plot (Figure 1A,1B). Then, the DEGs common to both data sets were identified via a Venn diagram. A comparison of HCC tissues to normal tissue yielded 132 common DEGs, of which 34 were upregulated (P value <0.05 and log2FC ≥1) and 98 were downregulated (P value <0.05 and log2FC ≤−1) in HCC tissues (Figure 1C).
Table 3
| Data source | Platform | Year | Sample size (tumor/normal) | Type | Genes (up/down) |
|---|---|---|---|---|---|
| GSE101728 | GPL21047 | 2019 | 7/7 | mRNA | 1,961/2,374 |
| GSE88839 | GPL570 | 2019 | 35/3 | mRNA | 108/265 |
mRNA, messenger RNA.
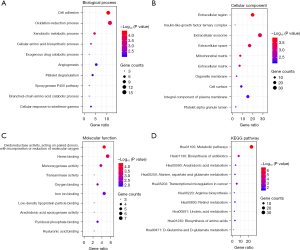
GO and KEGG pathway analysis of DEGs in HCC
The GO and KEGG pathway analysis of 132 DEGs was performed with DAVID database. The top 10 GO terms in the BP, CC, and MF categories are shown in Figure 2A-2C. The overlapping DEGs were mainly enriched in cell adhesion among the BP categories; the extracellular region in the CC categories; and oxidoreductase activity, acting on paired donors, and incorporation or reduction of molecular oxygen among the MF categories. The KEGG pathway analysis revealed that the DEGs were significantly enriched in the metabolic pathways (Figure 2D).
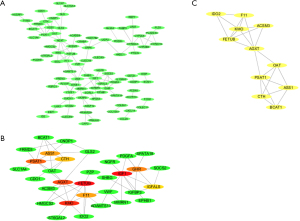
Analysis of PPI and modules
The 132 DEGs were assembled into a DEG PPI network complex comprising 118 edges and 132 nodes (PPI enrichment P value =1.0e–16; Figure 3A). Next, the top 10 hub genes were further analyzed using CytoHubba, which revealed that IGF1, FETUB, KMO, AGXT, PSAT1, GHR, F11, ASS1, CTH, and IGFALS were all downregulated genes (Figure 3B). Finally, according to the Cytoscape mCODE plug-in, the DEGs were enriched in the first clusters and with the score 3.833 (Figure 3C).
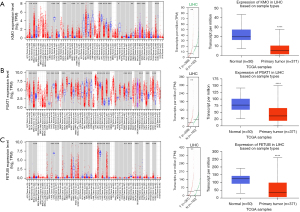
Analysis of hub genes via TIMER, UALCAN, and GEPIA
Through PubMed literature search, we finally selected KMO, FETUB, and PSAT1 as the key research biomarkers. TIMER, GEPIA, and UALCAN were applied to analyze the expression data for the 2 hub genes. The results showed that 3 genes (KMO, FETUB, and PSAT1) were underexpressed in HCC samples compared with normal liver samples (Figure 4), which was consistent with the results of the chip array and suggested a correlation with the occurrence of HCC.
Prediction of miRNAs that regulate key hub genes
The miRNAs that regulate KMO, FETUB, or PSAT1 were screened using TargetScan, starBase, TarBase, and the miRDB website. The results are shown in the Table 4 and Figure 5 (FETUB did not meet the threshold setting).
Table 4
| Gene symbol | Regulation | Target miRNAs |
|---|---|---|
| KMO | Down | hsa-miR-3613-5p |
| hsa-miR-6793-3p | ||
| hsa-miR-302f | ||
| hsa-miR-302c-5p | ||
| PSAT1 | Down | hsa-miR-15a-5p |
| hsa-miR-16-5p | ||
| hsa-miR-15b-5p | ||
| hsa-miR-195-5p | ||
| hsa-miR-424-5p | ||
| hsa-miR-429 | ||
| hsa-miR-497-5p | ||
| hsa-miR-340-5p | ||
| hsa-miR-224-5p | ||
| hsa-miR-18a-5p | ||
| hsa-miR-26a-5p | ||
| hsa-miR-26b-5p | ||
| hsa-miR-1-3p | ||
| hsa-miR-186-5p | ||
| hsa-miR-18b-5p | ||
| hsa-miR-485-3p | ||
| hsa-miR-1277-5p | ||
| hsa-miR-126-5p | ||
| hsa-miR-5680 | ||
| hsa-miR-570-3p | ||
| hsa-miR-200b-3p | ||
| hsa-miR-142-5p | ||
| hsa-miR-145-5p | ||
| hsa-miR-200c-3p | ||
| hsa-miR-323a-3p | ||
| hsa-miR-409-3p | ||
| hsa-miR-524-5p | ||
| hsa-miR-520d-5p | ||
| hsa-miR-545-3p | ||
| hsa-miR-577 | ||
| hsa-miR-579-3p | ||
| hsa-miR-1297 | ||
| hsa-miR-4524a-5p | ||
| hsa-miR-5094 | ||
| hsa-miR-5195-3p | ||
| hsa-miR-4524b-5p | ||
| hsa-miR-664b-3p | ||
| hsa-miR-5590-3p | ||
| hsa-miR-6838-5p |
miRNA, microRNA; KMO, kynurenine 3-monooxygenase.
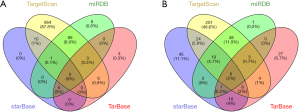
A total of 4 target miRNAs of KMO and 39 target miRNAs of PSAT1 were obtained from the online website (TarfetScan, miRDB, starBase & TarBase) (Figure 5A,5B).
Furthermore, we searched the PubMed database and selected the KMO and PSAT1 as candidate research molecules (Table 5). KMO had 4 potential target miRNAs and PSAT1 had 14 potential target miRNAs.
Table 5
| Gene symbol | Regulation | Target miRNAs |
|---|---|---|
| KMO | Down | hsa-miR-3613-5p |
| hsa-miR-6793-3p | ||
| hsa-miR-302f | ||
| hsa-miR-302c-5p | ||
| PSAT1 | Down | hsa-miR-1277-5p |
| hsa-miR-5680 | ||
| hsa-miR-570-3p | ||
| hsa-miR-200c-3p | ||
| hsa-miR-323a-3p | ||
| hsa-miR-409-3p | ||
| hsa-miR-524-5p | ||
| hsa-miR-579-3p | ||
| hsa-miR-1297 | ||
| hsa-miR-4524a-5p | ||
| hsa-miR-5094 | ||
| hsa-miR-5195-3p | ||
| hsa-miR-4524b-5p | ||
| hsa-miR-6838-5p |
miRNA, microRNA; KMO, kynurenine 3-monooxygenase.
Survival analysis
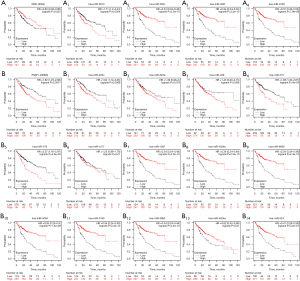
In order to further verify the effect of the candidate genes on HCC, the K-M plotter was applied to analyze their association with OS of the hub genes (Figure 6). We found that low KMO, hsa-miR-302c-5p, hsa-miR-302f, and hsa-miR-6793-3p expression was associated with a shorter OS in patients with HCC (Figure 6A,6A2-A4; P<0.05 and P<0.001), while, hsa-miR-3613-5p was associated with the longer OS of patients with HCC (Figure 6A1; P<0.01). Therefore, hsa-miR-3613-5p was chosen as the potential target miRNA of KMO. We subsequently found that the high expression of PSAT1, hsa-miR-200c-3p, hsa-miR-570-3p, and hsa-miR-579-3p was associated with the shorter OS of patients with HCC (Figure 6B1,B4-5; P<0.01 and P<0.001), while the low expression of hsa-miR-1297, hsa-miR-4524b-5p, hsa-miR-6838-5p, hsa-miR-5094, hsa-miR-5195-3p, hsa-miR-5680, hsa-miR-4524a-5p, and hsa-miR-524-5p was associated with the longer OS of patients with HCC (Figure 6B7-14; P<0.001, P<0.05, and P<0.001). The other miRNAs, hsa-miR-323a-3p, hsa-miR-409-3p, and hsa-miR-1277-5p did not show a statistically significant difference (Figure 6B2-3,B6; P>0.05). Therefore, hsa-miR-1297, hsa-miR-4524b-5p, hsa-miR-6838-5p, hsa-miR-5094, hsa-miR-5195-3p, hsa-miR-5680, hsa-miR-4524a-5p, and hsa-miR-524-5p were identified as candidate miRNAs target genes of PSAT1. These results further revealed that hsa-miR-3613-KMO, hsa-miR-1297, hsa-miR-4524b-5p, hsa-miR-6838-5p/hsa-miR-5094, hsa-miR-5195-3p, hsa-miR-5680, hsa-miR-4524a-5p, and hsa-miR-524-5p-PSAT1 are associated with the prognosis of HCC. To further analyze the role of these KMO and hsa-miR-3613 in HCC, we examined hsa-miR-3613-KMO in subsequent experiments.
KMO was downregulated in HCC cells
To explore the expression level of KMO in liver cancer cells, the normal liver cell line HL7702 and 4 liver cancer cell lines MHCC97, MHCC97-H, HCC-9903, and BEL-7402 were selected for qRT-PCR and Western blot experiments. The results showed that compared with normal liver cells, 4 liver cancer cell lines had significantly reduced expression levels of KMO mRNA and protein (Figure 7A,7B). It was concluded that KMO had low expression in liver cancer cells. It was also found that among these 4 liver cancer cell lines, KMO had the highest expression in MHCC97 and the lowest expression in MHCC97-H. Therefore, we chose MHCC97 and MHC997-H for follow-up experiments.

The expression level of KMO regulated the proliferation, invasion, migration, and EMT of HCC cells, and induced cell apoptosis
A KMO interfering RNA plasmid was transfected in MHCC97 cells, and a KMO-overexpressing plasmid was transfected in MHCC97-H cells. The qRT-PCR and Western blot results showed that the expression level of KMO in MHCC97 cells was significantly reduced after transfection of KMO interfering RNA plasmid but significantly increased in MHCC97-H cells after transfection of KMO-overexpressing plasmid (Figure 8A,8B). Subsequently, we examined the effects of KMO expression on the proliferation, invasion, migration, EMT, and apoptosis of HCC cells.
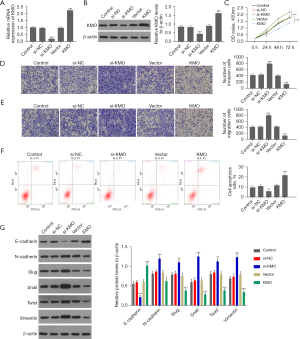
According to CCK-8 assay, knockdown of KMO significantly enhanced the viability of MHCC97 cells, while increased KMO expression level significantly inhibited the viability of MHCC97-H (Figure 8C). Transwell invasion and migration experiments showed that the decrease of KMO expression level could promote the invasion and migration ability of liver cancer MHCC97 cells, and the increase of expression level could inhibit the invasion and migration ability of liver cancer MHCC97-H cells (Figure 8D,8E). Flow cytometry detection of cell apoptosis showed that knocking down the expression of KMO could inhibit the apoptosis of liver cancer MHCC97 cells, while overexpression of KMO promoted the apoptosis of liver cancer MHCC97-H cells (Figure 8F). Western blot results showed that knocking down KMO could promote the expression of N-cadherin, slug, snail, twist, and vimentin, while inhibiting the expression of E-cadherin in MHCC97 cells; conversely, in MHCC97-H cells, the overexpression of KMO could inhibit N-cadherin, slug, snail, twist, and vimentin, while promoting the expression of E-cadherin (Figure 8G).
hsa-miR-3613-5p was highly expressed in HCC cells and negatively regulated the expression level of KMO
To verify the expression level hsa-miR-3613-5p in liver cancer cells, the normal liver cell line HL7702 and 4 liver cancer cell lines MHCC97, MHCC97-H, HCC-9903, and Bel-7402 were examined with qRT-PCR assay. The results showed that the expression level of hsa-miR-3613-5p was significantly increased in all 4 HCC cell lines compared with normal liver cells (Figure 9A). Among the 4 HCC lines, the expression of hsa-miR-3613-5p was the lowest in MHCC97 and the highest in MHCC97-H; we thus selected 2 cell lines, MHCC97 and MHC997-H, for subsequent experiments.

qRT-PCR results showed that the expression level of hsa-miR-3613-5p was significantly decreased while the expression level of KMO was significantly increased after knockdown of hsa-miR-3613-5p in MHCC97-H cells (Figure 9B,9C). After overexpression of hsa-miR-3613-5p, the expression level of hsa-miR-3613-5p was significantly increased and the expression level of KMO was significantly decreased in MHCC97 cells (Figure 9B,9C). These results indicated that hsa-miR-3613-5p is highly expressed in HCC cells and can negatively regulate the expression level of KMO.
Discussion
HCC is one of the major global public health problems and a leading cause of death. HCC is characterized by its rapid progression, rapid recurrence, rapid metastasis, high degree of malignancy, and high mortality (28,29). There are many treatments for HCC, such as radiotherapy, embolization, and chemotherapy (2,30). However, HCC patients have a high frequency of tumor recurrence after these treatments (31,32). In recent years, genome-wide expression profiling screening has attracted great attention in the diagnosis and treatment of patients with HCC (33). Thus, it is necessary to identify suitable molecular biomarkers for early diagnosis and potential targets for HCC therapies.
In this study, we found 132 integrated DEGs in HCC with a comprehensive analysis of the GSE101728 and GSE88839 data sets. The 132 integrated DEGs were then subjected to GO (BP, CC, and MF) analysis. The top 3 terms generated by the DEG enrichment analysis were the following: cell adhesion, reduction process, and xenobiotic metabolic process (BP); extracellular region, insulin-like growth factor ternary complex, and extracellular exosome (CC); and oxidoreductase activity, acting on paired donors, and incorporation or reduction of molecular oxygen, heme binding, and monooxygenase activity (MF). These results indicate that these DEGs are involved in the metabolic process, invasion, and metastasis of liver cancer cells. KEGG pathway analysis showed that DEGs were mainly enriched in metabolic pathways, biosynthesis of antibiotics, and arachidonic acid metabolism. Two different pathway enrichment metabolic processes showed that DEGs of HCC are involved in the process of cell metabolism. The alterations in intracellular and extracellular metabolites that can accompany cancer-associated metabolic reprogramming have profound effects on gene expression, cellular differentiation, and the tumor microenvironment (34). Thus, studying these pathways will help elucidate the underlying mechanisms of HCC proliferation and invasion and help predict cancer progression.
We constructed a PPI network with 132 integrated DEGs and identified the following 10 hub genes: IGF1, FETUB, KMO, AGXT, PSAT1, GHR, F11, ASS1, CTH, and IGFALS. Most of these factors affect the occurrence and development of cancer mainly by affecting the metabolic process. These central genes may be used as therapeutic targets for HCC. KMO, FETUB, and PSAT1 were analyzed in depth through PubMed literature research. We then performed an expression analysis of the 3 hub genes using the TIMER, GEPIA, and UALCAN websites. KMO and PSAT1 both had low expression in cancer tissue, consistent with the GEO data results, while FETUB not.
To further investigate KMO and PSAT1 and their function in the HCC, we predicted their miRNAs using the miRDB, TarBase v. 8.0, starBase v. 3.0, and TargetScan databases. Further analyses provided insight into the potential functions of these predicted associated miRNAs, especially those involved in the PI3K-Akt/AKT-MAPK signaling pathway (35,36). These results will help us understand the role of these candidate genes and provide potential biomarkers and targets for further clinical application in HCC prognostic monitoring and targeted therapy. In addition, we investigated the association of KMO-miRNAs and PSAT1-miRNAs with HCC prognosis. Surprisingly, the expression levels of hsa-miR-3613-5p-KMO, hsa-miR-1297/hsa-miR-4524b-5p, hsa-miR-6838-5p, hsa-miR-5094, hsa-miR-5195-3p, hsa-miR-5680, hsa-miR-4524a-5p, and hsa-miR-524-5p-PSAT1 were associated with the prognosis of patients with HCC cancer. Comprehensive literature analysis revealed hsa-miR-3613-5p-KMO to be a major pathway in HCC progression.
Previous research suggests that KMO exerts tumor-promoting effects in HCC 11, KMO expression levels were reported to be high in TNBC, and KMO knockdown was found to decrease lung metastasis and prolong survival (37); moreover, patients with colorectal cancer and higher KMO expression were associated with higher metastasis and poorer survival rates (38). In our study, KMO knockdown promoted the proliferation, invasion, metastasis, apoptosis, and EMT of HCC cells in vitro. Furthermore, using bioinformatics comprehensive analysis, we found that KMO may be the target molecule of miR-3613-5p. The role of miR-3613-5p has been investigated in previous studies. For example, Cao et al. demonstrated that miR-3613-5p enhances the metastasis of pancreatic cancer (39). He et al. reported that miR-3613-5p promotes the proliferation of lung adenocarcinoma (36). Qin et al. identified miR-3613-5p as a predictor of OS in HCC (40). In our study, we found that the expression of miR-361-3p was obviously increased in HCC cells as predicted by bioinformatics and PCR assays. Furthermore, miR-3613-5p knockdown stimulated the expression of KMO, while miR-3613-5p overexpression inhibited the expression of KMO. These results indicate that KMO may be a novel candidate molecular for human HCC.
However, there were several limitations in the present study. First, no further experimental validation for verifying the roles of miR-3613-5p was conducted; thus, extended functional studies are needed to investigate the roles of the miR-3613-5p in HCC cells. Second, experiments clarifying mechanisms of KMO in vivo should be conducted to complement the functional research of KMO in HCC. Despite these limitations, this study yielded an important finding, demonstrating that KMO may serve as a promising predictor of the OS in patients with HCC.
Conclusions
We conducted a comprehensive study in which the molecular biomarkers in patients with HCC were screened. We found that KMO was significantly underexpressed in HCC tissues and cells. Furthermore, KMO as an oncogene could promote HCC cell proliferation, migration, invasion, apoptosis, and EMT. These findings point to KMO as a candidate gene in HCC development and progression.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-147/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-147/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-147/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-147/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Vogel A, Meyer T, Sapisochin G, et al. Hepatocellular carcinoma. Lancet 2022;400:1345-62. [Crossref] [PubMed]
- Gilles H, Garbutt T, Landrum J. Hepatocellular Carcinoma. Crit Care Nurs Clin North Am 2022;34:289-301. [Crossref] [PubMed]
- Ganesan P, Kulik LM. Hepatocellular Carcinoma: New Developments. Clin Liver Dis 2023;27:85-102. [Crossref] [PubMed]
- Vogel A, Martinelli EESMO Guidelines Committee. Updated treatment recommendations for hepatocellular carcinoma (HCC) from the ESMO Clinical Practice Guidelines. Ann Oncol 2021;32:801-5. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Park SY, Nam JS. Kynurenine pathway enzyme KMO in cancer progression: A tip of the Iceberg. EBioMedicine 2020;55:102762. [Crossref] [PubMed]
- Zwilling D, Huang SY, Sathyasaikumar KV, et al. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell 2011;145:863-74. [Crossref] [PubMed]
- Thevandavakkam MA, Schwarcz R, Muchowski PJ, et al. Targeting kynurenine 3-monooxygenase (KMO): implications for therapy in Huntington's disease. CNS Neurol Disord Drug Targets 2010;9:791-800. [Crossref] [PubMed]
- Platten M, Nollen EAA, Röhrig UF, et al. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov 2019;18:379-401. [Crossref] [PubMed]
- Jin H, Zhang Y, You H, et al. Prognostic significance of kynurenine 3-monooxygenase and effects on proliferation, migration, and invasion of human hepatocellular carcinoma. Sci Rep 2015;5:10466. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Wixon J, Kell D. The Kyoto encyclopedia of genes and genomes--KEGG. Yeast 2000;17:48-55. [PubMed]
- Jiao X, Sherman BT. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012;28:1805-6. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020;48:D127-31. [Crossref] [PubMed]
- Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 2018;46:D296-302. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [Crossref] [PubMed]
- Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Jiang W, Yang W, Liu J, et al. Cancer-suppressing miR-520-3p gene inhibits proliferation, migration, and invasion of gastric cancer cells through targeted regulation of KLF7. Bull Cancer 2022;109:631-41. [Crossref] [PubMed]
- Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res 2021;149:1-61. [Crossref] [PubMed]
- Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol 2019;12:133. [Crossref] [PubMed]
- Renne SL, Sarcognato S, Sacchi D, et al. Hepatocellular carcinoma: a clinical and pathological overview. Pathologica 2021;113:203-17. [Crossref] [PubMed]
- Tsilimigras DI, Bagante F, Moris D, et al. Recurrence Patterns and Outcomes after Resection of Hepatocellular Carcinoma within and beyond the Barcelona Clinic Liver Cancer Criteria. Ann Surg Oncol 2020;27:2321-31. [Crossref] [PubMed]
- Sugawara Y, Hibi T. Surgical treatment of hepatocellular carcinoma. Biosci Trends 2021;15:138-41. [Crossref] [PubMed]
- Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 2007;45:938-47. [Crossref] [PubMed]
- Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: Still emerging. Cell Metab 2022;34:355-77. [Crossref] [PubMed]
- Yin H, Cui X. Knockdown of circHIPK3 Facilitates Temozolomide Sensitivity in Glioma by Regulating Cellular Behaviors Through miR-524-5p/KIF2A-Mediated PI3K/AKT Pathway. Cancer Biother Radiopharm 2021;36:556-67. [Crossref] [PubMed]
- He T, Shen H, Wang S, et al. MicroRNA-3613-5p Promotes Lung Adenocarcinoma Cell Proliferation through a RELA and AKT/MAPK Positive Feedback Loop. Mol Ther Nucleic Acids 2020;22:572-83. [Crossref] [PubMed]
- Huang TT, Tseng LM, Chen JL, et al. Kynurenine 3-monooxygenase upregulates pluripotent genes through beta-catenin and promotes triple-negative breast cancer progression. EBioMedicine 2020;54:102717. [Crossref] [PubMed]
- Liu CY, Huang TT, Chen JL, et al. Significance of Kynurenine 3-Monooxygenase Expression in Colorectal Cancer. Front Oncol 2021;11:620361. [Crossref] [PubMed]
- Cao R, Wang K, Long M, et al. miR-3613-5p enhances the metastasis of pancreatic cancer by targeting CDK6. Cell Cycle 2020;19:3086-95. [Crossref] [PubMed]
- Qin L, Huang J, Wang G, et al. Integrated analysis of clinical significance and functional involvement of microRNAs in hepatocellular carcinoma. J Cell Physiol 2019;234:23581-95. [Crossref] [PubMed]
(English Language Editor: J. Gray)


