
Performance of a blood-based RNA signature for gemcitabine-based treatment in metastatic pancreatic adenocarcinoma
Highlight box
Key findings
• We confirmed the robustness of the GemciTest, a blood-based signature to predict the efficacy of a gemcitabine-based regimen in first-line treatment for mPDAC patients.
What is known and what is new?
• There is no routine biomarker to predict efficacy of gemcitabine-based treatment in metastatic pancreatic adenocarcinoma. Liquid biopsies have a growing potential to be correlated with treatment efficacy in precision medicine.
• We revealed the potential of a blood-based RNA signature contributing to personalized therapy in the context of mPDAC.
What is the implication, and what should change now?
• Predictive tests such as the GemciTest may help clinicians to optimize the benefit/risk ratio for a gemcitabine-based regimen as first-line therapy in metastatic pancreatic adenocarcinoma.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) incidence is becoming more prevalent in Western countries, and it is projected to become the second most common cause of cancer-related deaths by 2030 (1). Despite its high prevalence and incidence rates, the prognosis for this disease remains grim, with an overall 5-year survival rate of under 10%. More than half of patients have metastases at diagnosis, and therapeutic options remain limited despite recent progress. Until 2011, the standard of care was gemcitabine; next came the FOLFIRINOX chemotherapy regimen [5-fluorouracil (5FU), leucovorin, irinotecan, oxaliplatin], and in 2013, the gemcitabine plus nab-paclitaxel regimen was introduced. These two multi-drug treatments significantly improved the median overall survival (OS) from 6.7 to respectively 11.1 and 8.5 months (2,3). Unfortunately, no large clinical trials have compared FOLFIRINOX directly to gemcitabine plus nab-paclitaxel as the first-line treatment for metastatic PDAC (mPDAC), thus complicating the optimal chemotherapy choice. A prospective neoadjuvant phase 2 trial in patients with resectable PDAC reported a globally similar benefit and tolerance to the two regimens (4).
The median OS of patients with mPDAC remains less than one year in recent phase 3 trials (5). The current gold-standard serum tumor marker is CA19-9, which is clinically used for disease monitoring. However, this marker has a poor specificity, impacting its sensitivity (6). As such, in the absence of emerging novel therapeutics for most mPDACs, an option to increase the efficacy of available therapies is to optimize their therapeutic use through the development of predictive biomarkers. Such predictive biomarkers could facilitate guiding clinicians towards the optimal choice of first-line therapy and increase their efficacy.
Over the last decade, an increasing number of translational research has been dedicated to identifying potential biomarkers for predicting prognosis and treatment response in PDAC. Most of them have been DNA- and tissue-based to evaluate potentially actionable mutations (7). The Pancreas Cancer Olaparib Ongoing (POLO) trial was a large phase 3 trial to evaluate the efficacy of maintenance therapy with olaparib in patients with a germline BRCA mutation and with a control disease under first-line platinum-based chemotherapy (2). Of the 3,315 patients who underwent screening, only 154 (~4.6%) were identified as gBRCA-mutated. This finding is an encouraging first step towards personalized treatment and companion diagnostics in PDAC treatment (8,9). In addition, a more recent preclinical approach is emerging in precision medicine with biopsy-derived pancreatic organoids (BDPOs) (10). However, in mPDAC, diagnosis is most often made through endoscopic ultrasound-guided fine-needle aspiration or liver biopsy. This point can be an obstacle to obtaining a good quality and quantity of tumor samples. All oncology societies have highlighted these concerns (11,12).
Among liquid biopsies, circulating tumor DNA (ctDNA) has a strong prognostic value, and its dynamic evolution under chemotherapy is correlated with treatment efficacy (13-15). There is no consensual methodology [digital polymerase chain reaction (PCR), next generation sequencing (NGS)], defined target (KRAS mutation, FGFR fusion, gene promoter methylation), or cut-off of mutant allelic frequency to detect it. In addition, expressed genes (mRNA, coding, and non-coding) in blood cells, circulating miRNA, and ctDNA are considered to be targetable biomarkers, either being a mirror of the known metabolic pathways in solid tumors or being shed by dying tumor cells into the circulation. Their analysis enables the detection of molecular alterations, strengthening our belief in their use as blood-based biomarkers. Thus, several blood biomarkers have been evaluated, such as circulating tumor cells, ctDNA, miRNA, and exosomes (16), as well as a blood-based predictive signatures (17), but only a few studies with a limited number of patients have compared these different biomarkers, and none of them are currently used in routine clinical practice.
Our objective was to investigate a blood-based RNA signature that can predict the efficacy of chemotherapy for first-line treatment in mPDAC. Although gemcitabine, either alone or in combination with nab-paclitaxel, has been shown to improve survival rates, response rates are often incomplete and nearly all tumors develop some degree of gemcitabine resistance (2,3). Indeed, previous clinical studies, after close-out, have found that between 77% and 93% of patients prescribed first-line gemcitabine-based therapy do not respond (2,3). Additionally, gemcitabine has many untoward side effects, which are well-described and often neglected (18). Finally, second-line treatment may not be possible due to a decline in the performance status or other measures of suitability for further chemotherapy. In first-line clinical trials, only half or less of patients can receive a second-line chemotherapy regimen (2,3).
In 2020, we developed an innovative blood-based RNA signature to predict gemcitabine-based efficacy in patients suffering from mPDAC (19). This test, called the GemciTest, is a non-invasive test based on a nine-gene blood-based RNA signature. In this validation study, we strengthened the predictive test value of this blood-based RNA signature to identify patients with a clinical response [progression-free survival (PFS) ≥3.5 months; OS >8.7 months] to a gemcitabine-based regimen in the first-line treatment of mPDAC. We present the following article in accordance with the REMARK reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-946/rc).
Methods
Patients and sample collection
A total of 336 blood samples were collected in PAXgene Blood RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland) from two distinct clinical studies (ClinicalTrials.gov: NCT00789633, NCT03599154) and two prospective cohorts (University of Wisconsin Carbone Cancer Center TSB-BioBank and BACAP-NCT02818829). The blood samples were collected during baseline blood work, before chemotherapy initiation. These cohorts included patients who had not yet received treatment, with either a gemcitabine- or fluoropyrimidine-based regimen. Gemcitabine, gemcitabine plus nab-paclitaxel, FOLFIRINOX (5FU, oxaliplatin, irinotecan), and FOLFOX (5FU, oxaliplatin) were administered according to standard clinical practice. For the subjects in clinical trials, informed consent was obtained from all subjects involved in the study. Treatments were administered until progression, intolerance, or patient withdrawal, with disease progression assessed via computed tomography (CT) scan according to the RECIST criteria every 8 weeks. In the event of a treatment-related grade 3 or 4 adverse event (AE), treatment interruption or blinded dose reduction was permitted according to the predefined criteria. The study was carried out in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the national health authorities and local ethics committees (ClinicalTrials.gov: NCT00789633, NCT03599154, University of Wisconsin Carbone Cancer Center TSB-BioBank and BACAP-NCT02818829). Blood samples were collected prior to the initiation of the treatment. For the subjects from the BioBanks, treatment and disease assessment were performed according to the institutional standard of care and treating physician preference. The main socio-demographic, clinical, biological, and histological data were collected and stored in an e-observation system at a centralized data center (20). According to the user’s manual for the PAXgene Blood RNA tubes, the tubes were inverted 10 times immediately after blood collection and incubated for a minimum of 2 hours at room temperature before freezing. Each tube contained 2.5 mL of whole blood and 6.9 mL of additives that prevent RNA degradation. The tubes were stored at −80 ℃ until use.
Gene expression analysis via quantitative PCR (qPCR)
The total RNA from the blood samples was extracted using a PAXgene Blood RNA Kit V2 (PreAnalytiX) on a QIAcube liquid handling platform according to the manufacturer’s protocols. The RNA purity and quantity were controlled using a Thermo Scientific NanoDrop ND-1000 spectrophotometer, and the RNA integrity was controlled with an Agilent 2200 TapeStation. The following quality requirements were applied: a minimum of 300 ng of total RNA with an absorbance ratio (260/280 nm) of >1.8. The gene expression analyses were performed using a LightCycler® 480 SYBR Green I Master (Roche Diagnostics, Santa Clara, CA, USA) in a 10 µL final reaction volume, according to the manufacturer’s protocol, using a LightCycler® 480 System II Instrument (Roche Diagnostics) and an established qPCR assay (19). The targeted genes were ATP-binding cassette subfamily C member 1 (ABCC1), ADP Ribosylation Factor-Like GTPase 4C (ARL4C), LYN proto-oncogene, Src family tyrosine kinase (LYN), NME/NM23 nucleoside diphosphate kinase 4 (NME4), peptidylprolyl isomerase B (PPIB), ubiquitin-conjugating enzyme E2 H (UBE2H), and transporters such as solute carrier family 35 member E2B (SLC35E2B). The normalization of gene expression levels was done using two housekeeping genes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and beta-2-microglobulin (B2M). We defined the acceptance criteria for real-time PCR analysis (19), including five replicate measures for all genes in compliance with the defined limit of quantification (LoQ), the melting temperature (Tm), and negative controls.
Statistical analysis: gene expression-based (GE) score
The GE score was previously established (19) with the exact methodology described in previous article (19). We then used signature built upon two components to predict the OS (Comp1) and PFS (Comp2) and used it to compute prediction. The combined result of these two components allows the GemciTest to determine if a patient is GemciTest positive or GemciTest negative. GemciTest positive is the condition where the usage of a gem-based treatment could be beneficial in terms of the PFS and OS for the patient.
Statistical analysis: univariate and multivariate analysis
Predictions from the Comp1 and Comp2 were added to the clinical covariates, and we performed both univariate and multivariate analysis testing using Cox proportional hazard models.
Statistical analysis: treatment-based cohort comparison
We performed a Fisher exact test or Chi-squared test for the comparison of qualitative variables between gemcitabine-based patients and 5FU-based patients; analysis of variance (ANOVA) was used for the comparison of quantitative variables.
Results
Following the current clinical recommendations (11,12,21), we separated the patients with mPDAC from those with locally advanced PDAC (LAPC). Indeed, LAPC is not a metastatic disease, although it is not amenable to radical surgery (22). From the four independent cohorts, and according to specific criteria, 142 of the 336 patients were considered for this validation phase (Figure 1): patients diagnosed with mPDAC, naïve of chemotherapy and treated in first-line with a gemcitabine- or 5FU-based regimen (Table 1). The median follow-up for alive patients was at least 12 months. According to the defined acceptance criteria for real-time PCR analysis, we excluded 17 samples because of a low extracted RNA quantity and one sample because of no PCR amplification for one gene. Patients who received gemcitabine plus nab-paclitaxel as first-line treatment represent 6% of gem-based patients, and patients treated with FOLFIRINOX as first-line treatment represent 88% of 5FU-based patients.
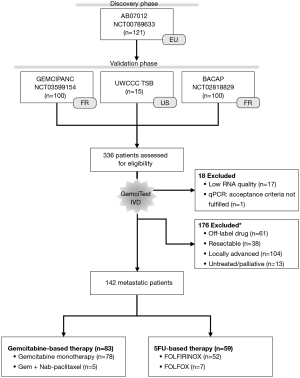
Table 1
| Baseline characteristics | All | GEM | 5FU | P value† |
|---|---|---|---|---|
| Number | 142 | 83 | 59 | |
| Gender (female) | 73 (51.4%) | 42 (50.6%) | 31 (52.5%) | 0.9541 |
| Age (years), median (range) | 68.0 (41.0–85.0) | 69.0 (41.0–85.0) | 66.0 (44.0–78.0) | 0.0472 |
| Body mass index (kg/m2), mean (SD) | 23.5 (4.1) | 23.8 (4.2) | 23.2 (4.1) | 0.487 |
| CA 19-9 (U/mL), mean (SD) | 34,783 [88,679] | 32,904 [96,349] | 38,540 [71,993] | 0.672 |
| Albumin (g/L), mean (SD) | 35.1 (6.4) | 34.4 (6.3) | 37.1 (6.5) | 0.65 |
| ECOG PS | 0.8406 | |||
| ECOG [0–1] | 102 (72%) | 61 (73%) | 41 (70%) | |
| ECOG [2+] | 34 (28%) | 19 (27%) | 15 (30%) | |
| Monocyte count (per µL), median (range) | 0.63 (0.0–1.85) | 0.61 (0.0–1.53) | 0.69 (0.38–1.85) | 0.344 |
| Tumor localization‡, n (%) | 0.1476 | |||
| Head | 58 (41%) | 40 (48%) | 18 (30%) | |
| Body | 53 (37%) | 31 (37%) | 22 (37%) | |
| Tail | 44 (31%) | 22 (26%) | 22 (37%) |
†, the Fisher exact test or Chi-squared test was used for comparison of qualitative variables between GEM and 5FU groups; analysis of variance was used for comparison of quantitative variables; ‡, patients presenting tumors in more than one location are included in both categories, BACAP cohort had missing data. Note: the sum of the percentages might not be equal to 100% or sum of patient might not be equal to their total if data were not available. PDAC, pancreatic ductal adenocarcinoma; 5FU, 5-fluorouracil; All, all patients included; GEM, patient subgroup treated with Gem-based chemotherapy as first line; 5FU, patient subgroup treated with 5FU-based chemotherapy; SD, standard deviation; CA19-9, carbohydrate antigen 19-9; ECOG PS, Eastern Cooperative Oncology Group Performance Status.
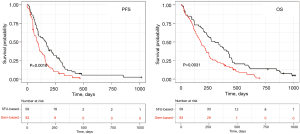
As expected from this real-world patient population, patients had an observed median OS and PFS of 5.8 and 2.8 months with gem-based regimens and of 10.1 and 5.6 months with 5FU-based regimens, respectively (Figure 2). Using Fisher exact test, Chi-squared test, and ANOVA, clinical characteristics of patients treated with gem-based versus 5FU-based were similar, except age with patients 5FU-based younger than patients gem-based (median 66 vs. 69 years; P=0.04) and a trend for a higher albuminemia level (Table 1).
Two groups of patients were defined according to the GemciTest: those with a positive test (GemciTest positive) and those with a negative test (GemciTest negative). The patients’ characteristics at baseline were fairly comparable between the two groups. In the subgroup of patients treated with a gemcitabine-based regimen (n=83), patients with a positive GemciTest (n=19/83; 22.9%) had a significantly longer PFS (5.3 vs. 2.8 months) and OS (10.4 vs. 4.8 months) (Figure 3A,3B, Table 2). In multivariate analyses, PFS {hazard ratio (HR) =0.53 [95% confidence interval (CI): 0.31–0.92]; P=0.023} and OS [HR =0.49 (95% CI: 0.29–0.85); P=0.0091] were significantly longer in patients with a positive GemciTest and treated with a gemcitabine-based regimen. In the subgroup of patients LAPC treated with a gem-based regimen (n=43), a univariate and multivariate cox model was fitted for both PFS and OS, for OS prediction the GemciTest was unable to stratify the patient (Comp1 and Comp2 P values were not significant), for PFS the Comp2 only is significant but still the GemciTest prediction is based on the two components Comp (Figure 3C,3D, Table S1). Also, in the subgroup of metastatic patients treated with a 5FU-based regimen (n=59), there was no significant difference in PFS and OS between the patients with a positive GemciTest (n=5/54; 9.2%) and the others (Figure 3E,3F, Table S2).
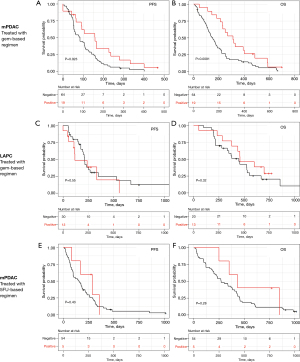
Table 2
| Baseline characteristics | Patients number | GemciTest classification | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | POS | PPFS | POS | PPFS | ||||
| Number | 83 | 64 | 19 | ||||||
| Comp1 | 83 | 2.03E–05 | 0.00746 | 0.11 | 9.12E–05 | ||||
| Comp2 | 83 | 0.165 | 0.148 | – | – | ||||
| Gender (female) | 41 (49%) | 29 (45%) | 12 (63%) | 0.85 | 0.852 | – | – | ||
| Age (years), median (range) | 69.5 (41.0–88.0) | 69.0 (41.0–88.0) | 70.0 (48.0–85.0) | 0.378 | 0.187 | – | – | ||
| Body mass index (kg/m2), mean (SD) | 23.8 (4.2) | 23.8 (4.2) | 23.7 (4.7) | 0.264 | 0.572 | – | – | ||
| CA 19-9 (U/mL), mean (SD) | 32,904 [96,349] | 42,340 [109,228] | 3,548 [7,612] | 0.162 | 0.619 | – | – | ||
| Albumin (g/L), mean (SD) | 34.4 (6.3) | 32.9 (6.1) | 38.6 (4.7) | 0.0101 | 0.239 | 0.5 | – | ||
| ECOG PS | 0.01 | 0.175 | 0.75 | – | |||||
| ECOG [0–1] | 61 (73%) | 45 (70%) | 16 (84%) | ||||||
| ECOG [2-4] | 20 (24%) | 18 (28%) | 2 (10%) | ||||||
| Monocyte count (per µL), median (range) | 0.61 (0.0–1.53) | 0.65 (0.0–1.53) | 0.53 (0.19–1.25) | 0.434 | 0.692 | – | – | ||
| Tumor localization‡, n (%) | |||||||||
| Head | 40 (48%) | 31 (48%) | 9 (47%) | 0.509 | 0.679 | – | – | ||
| Body | 31 (37%) | 23 (36%) | 8 (42%) | 0.87 | 0.657 | – | – | ||
| Tail | 22 (27%) | 18 (28%) | 4 (21%) | 0.709 | 0.0854 | – | – | ||
‡, patients presenting tumors in more than one location are included in both categories. Note: the sum of the percentages might not be equal to 100%, and the sum of patients might not be equal to their total if the data were not available. PDAC, pancreatic ductal adenocarcinoma; P, P value; OS, overall survival; PFS, progression-free survival; Comp1 and Comp2, gene-set component 1 and 2; SD, standard deviation; CA19-9, carbohydrate antigen 19-9; ECOG PS, Eastern Cooperative Oncology Group Performance Status.
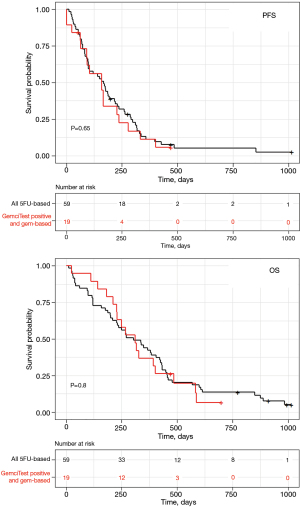
Patients treated with the FOLFIRINOX regimen had a longer OS than those treated with a gemcitabine-based regimen (Figure 2). Patients with a positive GemciTest and treated with a gemcitabine-based regimen had a similar median PFS (5.6 months for 5FU-based patients vs. 5.3 months for gem-based ones) and OS (10.1 vs. 10.4 months) than whole patients treated with a 5FU-based regimen (Figure 4); no difference for PFS [HR =1.1 (95% CI: 0.66–2); P=0.65] and OS [HR =1.1 (95% CI: 0.62–1.9); P=0.8] was shown in multivariate analyses.
Discussion
The nine targeted genes identified in a pre-discovery study from NGS-based transcriptomic analysis (23) are involved in chemotherapy responses and resistance mechanisms. For instance, it is noteworthy that targeted genes are crosslinked in lipid metabolism. Different studies identified lipid metabolism pathway significantly correlated with poor gemcitabine response in tumor tissues of patients with PDAC (24,25). Other mechanisms such as the human equilibrative nucleoside transporters (hENTs)/Solute Carrier Family deficiency have been described in gemcitabine resistance (24,25). Expression of the human equilibrative nucleoside transporter 1 (hENT1) and of the deoxycytidine kinase (dCK), two important proteins involved in the gemcitabine metabolism, have been suggested as potentially predictive biomarkers of its efficacy but no validated test is currently available to assess their expression in routine practice (26,27). While these classical “tumor cells” markers of gemcitabine sensitivity were not part of the blood-based gene expression signatures, one of the gene ABCC1, an efflux pump of the (ABC) transporter family proteins, was demonstrated to be involved in gemcitabine resistance (28). Indeed, chemo-resistance in PDAC patients is a challenging problem with scarce choices of chemotherapeutic agents. Although gemcitabine-based chemotherapy is one of the mainstays treatments for advanced mPDAC, clinical experience does not show a significant drug response to increase patient duration. Indeed, selected genes are known to be associated with gemcitabine resistance as the ABC transporter, ABCC1 (29,30), or the kinase NME4 (28), both included in the pyrimidine deoxyribonucleotides de novo biosynthesis pathway. Another mechanism is enlightened with ABCC1 and PPIB markers, playing a role in reactive oxygen species (ROS) production and control of redox metabolism, inducing ROS accumulation and activating Nrf2 signaling pathways (NRF2-mediated oxidative stress response) (31).
In this study, we confirm the robustness of the GemciTest (0.3% of sample were not evaluable) and the predictive ability of GemciTest to predict the efficacy of a gemcitabine-based regimen in first-line treatment for mPDAC patients. This study demonstrates the first exploratory ones, with a more significant number of patients from independent cohorts. The clinical variables for patients (metastatic status, performance status, albumin level, etc.) were included in the multivariate analyses to ensure that the value of the blood-based RNA signature was independent of them. In addition, we focused on mPDAC patients because recent works have shown that chemotherapy with FOLFIRINOX may have advantages relative to gemcitabine plus nab-paclitaxel and may be considered preferentially for patients without contraindications and who are anticipated to tolerate it [age of <75 years, Eastern Cooperative Oncology Group Performance Status (ECOG PS) 0–1].
Nevertheless, it is important to note that this study may have certain limitations. The estimates in our model were derived from prospective observational studies, which are susceptible to biases and confounding factors that could have influenced our findings. Also, the majority of the patients (94%) who received gemcitabine-based chemotherapy received gemcitabine monotherapy. We demonstrated that patient selection based on the GemciTest could be associated with a non-inferior oncologic outcome in comparison to a 5FU-based regimen, confirming its promising predictive value to choose the best chemotherapy regime for a given patient. The prospective validation of the GemciTest is currently ongoing in a randomized phase 3 trial, GEMFOX (NCT041667007), comparing gemcitabine monotherapy to FOLFOX in mPDAC in first-line patients unfit for FOLFIRINOX.
Conclusions
The GemciTest is based on real-world data, patient health status, and/or the delivery of routine health care, collected from four prospective cohorts (two clinical trials and two prospective cohorts). This blood-based RNA signature shows clinical evidence of optimizing the benefit/risk ratio for a gemcitabine-based regimen as first-line therapy in metastatic pancreatic adenocarcinoma. Although these data are promising, they should be confirmed in a prospective trial. Combining this test with dihydropyrimidine dehydrogenase (DPD) testing (either the enzyme activity of dihydropyrimidine dehydrogenase or the DPYD genotype) could represent the first step of precision medicine in mPDAC care. It must be emphasized that the blood sampling needed for the RNA signature analysis is performed on an outpatient basis. It can be carried out in a medical routine within 2 days and represents only a slight constraint for the patient, the hospital, and the nursing staff, allowing us to treat every patient as an exception. While waiting for new drugs is potentially only effective in rare molecular subgroups [KRAS G12C, BRCA1-2 germline mutation, microsatellite instability/deficient mismatch repair (MSI/dMMR), etc.] (32), optimizing patient selection for existing treatments is a promising strategy.
Acknowledgments
We thank the patients who participated in this trial and their families, as well as all the participating investigators and clinical staff. The authors thank the University of Wisconsin Carbone Cancer Center BioBank, supported by P30 CA014520 for the use of its facilities and services.
Funding: This work was supported by ACOBIOM.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-946/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-946/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-946/coif). DP, RB, FN, FP and A Gamez report that they are employed by Acobiom company. JBB reports that he received personal fees from Acobiom, Amgen, AstraZeneca, Bayer, Merck Serono, Pierre Fabre, Roche, Sanofi, Servier, Viatris, and non-financial support from Amgen, Merck Serono, and Roche. NKL reports that she is advisory board member for Abbvie and PDGX). The other authors have no conflicts of to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was carried out in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the national health authorities and local ethics committees (ClinicalTrials.gov: NCT00789633, NCT03599154, University of Wisconsin Carbone Cancer Center TSB-BioBank and BACAP-NCT02818829). Informed consent was obtained from all subjects involved in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Park W, Chawla A, O'Reilly EM. Pancreatic Cancer: A Review. JAMA 2021;326:851-62. [Crossref] [PubMed]
- Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 2013;369:1691-703. [Crossref] [PubMed]
- Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011;364:1817-25. [Crossref] [PubMed]
- Ahmad SA, Duong M, Sohal DPS, et al. Surgical Outcome Results From SWOG S1505: A Randomized Clinical Trial of mFOLFIRINOX Versus Gemcitabine/Nab-paclitaxel for Perioperative Treatment of Resectable Pancreatic Ductal Adenocarcinoma. Ann Surg 2020;272:481-6. [Crossref] [PubMed]
- Singh RR, O'Reilly EM. New Treatment Strategies for Metastatic Pancreatic Ductal Adenocarcinoma. Drugs 2020;80:647-69. [Crossref] [PubMed]
- Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol 2012;3:105-19. [PubMed]
- Lowery MA, Jordan EJ, Basturk O, et al. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clin Cancer Res 2017;23:6094-100. [Crossref] [PubMed]
- Golan T, Hammel P, Reni M, et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N Engl J Med 2019;381:317-27. [Crossref] [PubMed]
- Lai E, Ziranu P, Spanu D, et al. BRCA-mutant pancreatic ductal adenocarcinoma. Br J Cancer 2021;125:1321-32. [Crossref] [PubMed]
- Nicolle R, Gayet O, Bigonnet M, et al. Relevance of biopsy-derived pancreatic organoids in the development of efficient transcriptomic signatures to predict adjuvant chemosensitivity in pancreatic cancer. Transl Oncol 2022;16:101315. [Crossref] [PubMed]
- Philip PA, Mooney M, Jaffe D, et al. Consensus report of the national cancer institute clinical trials planning meeting on pancreas cancer treatment. J Clin Oncol 2009;27:5660-9. [Crossref] [PubMed]
- Van Laethem JL, Verslype C, Iovanna JL, et al. New strategies and designs in pancreatic cancer research: consensus guidelines report from a European expert panel. Ann Oncol 2012;23:570-6. [Crossref] [PubMed]
- Abdallah R, Taly V, Zhao S, et al. Plasma circulating tumor DNA in pancreatic adenocarcinoma for screening, diagnosis, prognosis, treatment and follow-up: A systematic review. Cancer Treat Rev 2020;87:102028. [Crossref] [PubMed]
- Bachet JB, Blons H, Hammel P, et al. Circulating Tumor DNA is Prognostic and Potentially Predictive of Eryaspase Efficacy in Second-line in Patients with Advanced Pancreatic Adenocarcinoma. Clin Cancer Res 2020;26:5208-16. [Crossref] [PubMed]
- Pietrasz D, Pécuchet N, Garlan F, et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin Cancer Res 2017;23:116-23. [Crossref] [PubMed]
- Pan Y, Li K, Tao X, et al. MicroRNAs in Pancreatic Cancer and Chemoresistance. Pancreas 2021;50:1334-42. [Crossref] [PubMed]
- Sakai Y, Honda M, Matsui S, et al. Development of novel diagnostic system for pancreatic cancer, including early stages, measuring mRNA of whole blood cells. Cancer Sci 2019;110:1364-88. [Crossref] [PubMed]
- Alam S, Illo C, Ma YT, et al. Gemcitabine-Induced Cardiotoxicity in Patients Receiving Adjuvant Chemotherapy for Pancreatic Cancer: A Case Series. Case Rep Oncol 2018;11:221-7. [Crossref] [PubMed]
- Piquemal D, Noguier F, Pierrat F, et al. Predictive Values of Blood-Based RNA Signatures for the Gemcitabine Response in Advanced Pancreatic Cancer. Cancers (Basel) 2020;12:3204. [Crossref] [PubMed]
- Canivet C, Gourgou-Bourgade S, Napoléon B, et al. A prospective clinical and biological database for pancreatic adenocarcinoma: the BACAP cohort. BMC Cancer 2018;18:986. [Crossref] [PubMed]
- Barros AG, Pulido CF, Machado M, et al. Treatment optimization of locally advanced and metastatic pancreatic cancer Int J Oncol 2021;59:110. (Review). [Crossref] [PubMed]
- Bosetti C, Bertuccio P, Malvezzi M, et al. Cancer mortality in Europe, 2005-2009, and an overview of trends since 1980. Ann Oncol 2013;24:2657-71. [Crossref] [PubMed]
- Deplanque G, Demarchi M, Hebbar M, et al. A randomized, placebo-controlled phase III trial of masitinib plus gemcitabine in the treatment of advanced pancreatic cancer. Ann Oncol 2015;26:1194-200. [Crossref] [PubMed]
- Sunami Y, Rebelo A, Kleeff J. Lipid Metabolism and Lipid Droplets in Pancreatic Cancer and Stellate Cells. Cancers (Basel) 2017;10:3. [Crossref] [PubMed]
- Tadros S, Shukla SK, King RJ, et al. De Novo Lipid Synthesis Facilitates Gemcitabine Resistance through Endoplasmic Reticulum Stress in Pancreatic Cancer. Cancer Res 2017;77:5503-17. [Crossref] [PubMed]
- Raffenne J, Nicolle R, Puleo F, et al. hENT1 Testing in Pancreatic Ductal Adenocarcinoma: Are We Ready? A Multimodal Evaluation of hENT1 Status. Cancers (Basel) 2019;11:1808. [Crossref] [PubMed]
- Maréchal R, Bachet JB, Mackey JR, et al. Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology 2012;143:664-674.e6. [Crossref] [PubMed]
- Phan T, Nguyen VH, Buettner R, et al. Inhibition of de novo pyrimidine synthesis augments Gemcitabine induced growth inhibition in an immunocompetent model of pancreatic cancer. Int J Biol Sci 2021;17:2240-51. [Crossref] [PubMed]
- Le Large TYS, El Hassouni B, Kazemier G, et al. Multidrug-resistant transporter expression does not always result in drug resistance. Cancer Sci 2018;109:3360-2. [Crossref] [PubMed]
- Wei L, Lin Q, Lu Y, et al. Cancer-associated fibroblasts-mediated ATF4 expression promotes malignancy and gemcitabine resistance in pancreatic cancer via the TGF-β1/SMAD2/3 pathway and ABCC1 transactivation. Cell Death Dis 2021;12:334. [Crossref] [PubMed]
- Abdel Hadi N, Reyes-Castellanos G, Carrier A. Targeting Redox Metabolism in Pancreatic Cancer. Int J Mol Sci 2021;22:1534. [Crossref] [PubMed]
- Principe DR. Precision Medicine for BRCA/PALB2-Mutated Pancreatic Cancer and Emerging Strategies to Improve Therapeutic Responses to PARP Inhibition. Cancers (Basel) 2022;14:897. [Crossref] [PubMed]


