
Network and pathway-based analysis of candidate genes associated with esophageal adenocarcinoma
Highlight box
Key findings
• We found that biochemical processes and pathways associated with the metabolic process, biological regulation process, and the signal transduction process played key roles in the molecular mechanism of esophageal adenocarcinoma (EA). In addition, a number of new genes were identified in the EA-specific network.
What is known and what is new?
• We know which genes and their pathways have been studied in EA.
• We found that biochemical processes and pathways associated with the metabolic process, biological regulation process, and the signal transduction process played key roles in the molecular mechanism of EA. In addition, there were 2 interconnected pathway modules: metabolic regulation and drug reactions, and transcriptional regulation. Finally, we extracted an EA-specific subnetwork and identified some novel genes potentially bound up with EA.
What is the implication, and what should change now?
• Our results suggested that in future studies in EA, we should focus on the pathways and new genes involved.
Introduction
Esophageal cancer, the ninth most common cancer and the sixth most fatal cancer worldwide, is classified into 2 major histologic types, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EA), the latter of which has increased rapidly in Western countries over the past several decades (1). Risk factors for EA include Barrett’s esophagus, gastroesophageal reflux disease, Caucasian race, male gender, obesity, smoking, and some genetic factors (2). EA has shown a poor prognosis, with an overall survival rate of approximately 20% at 5 years (3). Recently, with the development of neoadjuvant chemoradiotherapy and radiotherapy, survival rates have improved in patients with locally advanced EA compared to surgery alone, but there is wide interindividual variation in response to neoadjuvant therapy (4). In recent years, analysis of the genome of patients with EA has become increasingly relevant for guiding treatment planning.
The genomic complexity of EA is characterized by a high burden of point mutations and genome structural alterations, including TP53, CDKN2A, KRAS, MYC, and CDK6 (3). Rapid advances in high throughput technologies over the past decade have helped researchers generate numerous genetic and genomic datasets in order to reveal causal genes and their actions in complex diseases (5). Pathway analysis examines series of actions or interactions among genes or genes products that lead to the generation of a certain product or a change in the cell and protein-protein interaction (PPI) network, and it has been recognized as a powerful tool for understanding how genes perform their biological function (6).
In this study, we conducted a comprehensive analysis of genes which have been published potentially involved in EA. We then performed biological enrichment analyses and the interaction among the enriched biochemical pathways was analyzed, in addition to examining the crosstalk among the significantly enriched pathways. Finally, a molecular network of EA was constructed. We present the following article in accordance with the STREGA reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1286/rc).
Methods
Identification of EA-related genes
The genes genetically associated with EA (the standardized term found though MeSH) were gathered by retrieving human genetic association studies published in PubMed (http://www.ncbi.nlm.nih.gov/pubmed/). We retrieved 458 articles published to June 30, 2022 using the search terms (adenocarcinoma of esophageal and polymorphism [MeSH]) or (adenocarcinoma of esophageal and genotype [MeSH]) or (adenocarcinoma of esophageal and alleles [MeSH]), with ‘humans’ as the limiting condition. After browsing the abstracts of all of the articles, we excluded those which were not gene-related or were not focused primarily on EA, which left 125 articles remaining. We then focused on studies reporting a significant association between 1 or more genes with EA. For the purpose of reducing the number of potential false-positive genes, the studies reporting negative or insignificant associations were excluded even though some of them might influence EA in further research based on a large number of studies. Finally, we read the full text of each selected article to ensure the conclusion was in accord with its contents, and we also added some genes that were synergistic to the investigated genes. Eventually, we found 109 genes related to EA. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Function enrichment analysis
We used software from the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/) to convert the names of 109 EA genes from the literature into Entrez Gene GeneIDs. To examine the functional features of EA genes, Gene Ontology (GO, http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.kegg.jp/) were applied for functional enrichment analysis. In brief, GO enrichment analysis was used to annotate and classify the candidate genes of EA according to molecular function (MF), biological process (BP), and cellular component (CC). In addition, directed acyclic graph (DAG) was used to depict the results of GO enrichment analysis. In DAG, the top 10 terms with the lowest P value in the rectangle and their parent nodes in the circle were shown, with the branches indicating annotation moving from more general to more specific as one moved from parent nodes to child nodes, and the defined function range became smaller from top to bottom. The terms with pane marks indicated significant enrichment, with more red indicating more significance. We then applied KOBAS 2.1.1 software for comparison with the genes included in each pathway in the KEGG database, which is a knowledge base for systematic analysis of gene functions in order to link genomic information with higher order functional information. Next, we extracted the significantly enriched pathways and assigned a P value for each of them using Fisher’s exact test. In our study, both the GO and KEGG biological process terms with a multiple testing correction P value [false discovery rate (FDR)] <0.05 calculated using the Benjamini-Hochberg procedure were considered to be significantly enriched. In addition, we evaluated the degree of gene enrichment in a single function by enrichment score with the formula:
Pathway crosstalk analysis
We further performed pathway crosstalk analysis to investigate interactions of the enriched pathways. In this study, we used 2 measures to describe the overlap between any 2 pathways, the overlap coefficient (OC) and the Jaccard coefficient (JC).
where A and B are the lists of genes included in the 2 pathways under examination. The following procedure was carried out to construct the pathway crosstalk:
- We first selected a set of pathways for crosstalk analysis. Only the pathways with FDR <0.05 were kept. At the same time, more than 5 candidate genes were required in each pathway as pathways with too few genes might not have had sufficient biological information.
- Next, we counted the shared candidate genes of each pathway pair and removed pathways with fewer than 3 overlapping genes.
- We then calculated the JC and OC of the qualified pathway pairs and ranked them according to their score values.
Lastly, we visualized the final pathway crosstalk with Cytoscape software (7). The node size represented the degree of the pathways, with a larger node corresponding to a deeper degree, and the thickness of the line represented the score value of pathways, with a thicker line corresponding to a higher score.
Construction of the EA-specific protein subnetwork
Human PPI data was downloaded from the Protein Interaction Network Analysis (PINA) platform (8), which pools and curates unique physical interaction information from 6 main public PPI databases (IntAct, BioGRID, MINT, DIP, HPRD , and MIPS/MPact). We then used the Klein-Ravi algorithm in GenRev software to extract the subnetwork using the 109 EA genes as seeds (9). To examine the nonrandomness of the constructed network, 1000 random networks with the same number of nodes and edges as the EA-specific network were generated using the Erdos-Renyi model in the igraph package in R. Afterwards, we compared the seed genes with the 1000 random networks to generate subnetworks and calculated the average values of the shortest-path distance and clustering coefficient in random subnetworks. We estimated the significance of nonrandomness by counting the number of random networks with average shortest-path distance (nL) less than that of the EA-specific network and the number of random networks with average clustering coefficient (nc) more than that of the EA-specific network. Finally, we calculated the P-value = nL/1000 and nc/1000.
Results
Identification of genes related to EA
By searching PubMed, we collected literature on genetic associations related to EA. We selected 125 publications which reported gene(s) significantly associated with EA and collected 109 genes related to EA (Table 1). Some of the genes were involved in transcriptional regulation, such as hypoxia-inducible factor-1 (HIF-1) signaling (FLT1, BCL2, IGF1R, VEGFA, and PIK3CA), tumor necrosis factor (TNF) signaling (IL1B, MMP14, and PTGS2), vascular endothelial growth factor (VEGF) signaling (CASP9, KRAS), and others may be involved in drug metabolism, such as glutathione S-transferase mu 1 (GSTM1), glutathione S-transferase mu 3 (GSTM3), and cytochrome P450 family 2 subfamily E member 1 (CYP2E1).
Table 1
| Gene abbreviations | Gene ID | Species | Gene name |
|---|---|---|---|
| CTHRC1 | 115908 | Homo sapiens | Collagen triple helix repeat containing 1 (CTHRC1) |
| FHIT | 2272 | Homo sapiens | Fragile histidine triad (FHIT) |
| PTGS2 | 5743 | Homo sapiens | Prostaglandin-endoperoxide synthase 2 (PTGS2) |
| GDF7 | 151449 | Homo sapiens | Growth differentiation factor 7 (GDF7) |
| ASCC1 | 51008 | Homo sapiens | Activating signal cointegrator 1 complex subunit 1 (ASCC1) |
| SELENBP1 | 8991 | Homo sapiens | Selenium binding protein 1 (SELENBP1) |
| MMP3 | 4314 | Homo sapiens | Matrix metallopeptidase 3 (MMP3) |
| XRCC1 | 7515 | Homo sapiens | X-ray repair cross complementing 1 (XRCC1) |
| MMP2 | 4313 | Homo sapiens | Matrix metallopeptidase 2 (MMP2) |
| IL10 | 3586 | Homo sapiens | Interleukin 10 (IL10) |
| MMP1 | 4312 | Homo sapiens | Matrix metallopeptidase 1 (MMP1) |
| PGR | 5241 | Homo sapiens | Progesterone receptor (PGR) |
| GSTM1 | 2944 | Homo sapiens | Glutathione S-transferase mu 1 (GSTM1) |
| GSTM3 | 2947 | Homo sapiens | Glutathione S-transferase mu 3 (GSTM3) |
| MUTYH | 4595 | Homo sapiens | mutY DNA glycosylase (MUTYH) |
| CDKN2A | 1029 | Homo sapiens | cyclin dependent kinase inhibitor 2A (CDKN2A) |
| GATA6 | 2627 | Homo sapiens | GATA binding protein 6 (GATA6) |
| FOXF1 | 2294 | Homo sapiens | Forkhead box F1 (FOXF1) |
| GATA4 | 2626 | Homo sapiens | GATA binding protein 4 (GATA4) |
| IL1B | 3553 | Homo sapiens | Interleukin 1 beta (IL1B) |
| PIK3CA | 5290 | Homo sapiens | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) |
| NQO1 | 1728 | Homo sapiens | NAD (P)H quinone dehydrogenase 1 (NQO1) |
| WWOX | 51741 | Homo sapiens | WW domain containing oxidoreductase (WWOX) |
| EGFR | 1956 | Homo sapiens | Epidermal growth factor receptor (EGFR) |
| NEIL2 | 252969 | Homo sapiens | nei like DNA glycosylase 2 (NEIL2) |
| GSTT1 | 2952 | Homo sapiens | Glutathione S-transferase theta 1 (GSTT1) |
| ARID1A | 8289 | Homo sapiens | AT-rich interaction domain 1A (ARID1A) |
| CYP2E1 | 1571 | Homo sapiens | Cytochrome P450 family 2 subfamily E member 1 (CYP2E1) |
| CDO1 | 1036 | Homo sapiens | Cysteine dioxygenase type 1 (CDO1) |
| RFC3 | 5983 | Homo sapiens | Replication factor C subunit 3 (RFC3) |
| VEGFA | 7422 | Homo sapiens | Vascular endothelial growth factor A (VEGFA) |
| HPP1 | 780897 | Homo sapiens | Hyperpigmentation, progressive, 1 (HPP1) |
| FGFR2 | 2263 | Homo sapiens | Fibroblast growth factor receptor 2 (FGFR2) |
| WNT5A | 7474 | Homo sapiens | Wnt family member 5A (WNT5A) |
| MCL1 | 4170 | Homo sapiens | BCL2 family apoptosis regulator (MCL1) |
| CRTC1 | 23373 | Homo sapiens | CREB regulated transcription coactivator 1 (CRTC1) |
| ERBB2 | 2064 | Homo sapiens | erb-b2 receptor tyrosine kinase 2 (ERBB2) |
| TIMP3 | 7078 | Homo sapiens | TIMP metallopeptidase inhibitor 3 (TIMP3) |
| EPHB4 | 2050 | Homo sapiens | EPH receptor B4 (EPHB4) |
| WT1 | 7490 | Homo sapiens | Wilms tumor 1 (WT1) |
| VDR | 7421 | Homo sapiens | vitamin D (1,25- dihydroxyvitamin D3) receptor (VDR) |
| KRAS | 3845 | Homo sapiens | KRAS proto-oncogene, GTPase (KRAS) |
| DMD | 1756 | Homo sapiens | Dystrophin (DMD) |
| EGF | 1950 | Homo sapiens | Epidermal growth factor (EGF) |
| RUNX1 | 861 | Homo sapiens | Runt related transcription factor 1 (RUNX1) |
| RUNX3 | 864 | Homo sapiens | Runt related transcription factor 3 (RUNX3) |
| CYP19A1 | 1588 | Homo sapiens | Cytochrome P450 family 19 subfamily A member 1 (CYP19A1) |
| TP53BP1 | 7158 | Homo sapiens | Tumor protein p53 binding protein 1 (TP53BP1) |
| MET | 4233 | Homo sapiens | MET proto-oncogene, receptor tyrosine kinase (MET) |
| SMAD4 | 4089 | Homo sapiens | SMAD family member 4 (SMAD4) |
| FOXP1 | 27086 | Homo sapiens | Forkhead box P1 (FOXP1) |
| NR1I2 | 8856 | Homo sapiens | Nuclear receptor subfamily 1 group I member 2 (NR1I2) |
| FBXO32 | 114907 | Homo sapiens | F-box protein 32 (FBXO32) |
| PTPN1 | 5770 | Homo sapiens | Protein tyrosine phosphatase, non-receptor type 1 (PTPN1) |
| ABL1 | 25 | Homo sapiens | ABL proto-oncogene 1, non-receptor tyrosine kinase (ABL1) |
| IER3 | 8870 | Homo sapiens | Immediate early response 3 (IER3) |
| MSR1 | 4481 | Homo sapiens | Macrophage scavenger receptor 1 (MSR1) |
| CCNE1 | 898 | Homo sapiens | Cyclin E1 (CCNE1) |
| BARX1 | 56033 | Homo sapiens | BARX homeobox 1 (BARX1) |
| CASP9 | 842 | Homo sapiens | Caspase 9 (CASP9) |
| CASP7 | 840 | Homo sapiens | Caspase 7 (CASP7) |
| HMOX1 | 3162 | Homo sapiens | Heme oxygenase 1 (HMOX1) |
| CASP8 | 841 | Homo sapiens | Caspase 8 (CASP8) |
| NOS3 | 4846 | Homo sapiens | Nitric oxide synthase 3 (NOS3) |
| MYB | 4602 | Homo sapiens | MYB proto-oncogene, transcription factor (MYB) |
| MYC | 4609 | Homo sapiens | v-myc avian myelocytomatosis viral oncogene homolog (MYC) |
| TERT | 7015 | Homo sapiens | Telomerase reverse transcriptase (TERT) |
| TP53 | 7157 | Homo sapiens | Tumor protein p53 (TP53) |
| PRKCI | 5584 | Homo sapiens | Protein kinase C iota (PRKCI) |
| CDK6 | 1021 | Homo sapiens | Cyclin dependent kinase 6 (CDK6) |
| PADI4 | 23569 | Homo sapiens | Peptidyl arginine deiminase 4 (PADI4) |
| MBNL1 | 4154 | Homo sapiens | Muscleblind like splicing regulator 1 (MBNL1) |
| CDK4 | 1019 | Homo sapiens | Cyclin dependent kinase 4 (CDK4) |
| MMP14 | 4323 | Homo sapiens | Matrix metallopeptidase 14 (MMP14) |
| MMP12 | 4321 | Homo sapiens | Matrix metallopeptidase 12 (MMP12) |
| TNFRSF10A | 8797 | Homo sapiens | TNF receptor superfamily member 10a (TNFRSF10A) |
| VSIG10L | 147645 | Homo sapiens | V-set and immunoglobulin domain containing 10 like (VSIG10L) |
| Myo9B | 4650 | Homo sapiens | Myosin IXB (MYO9B) |
| NCOA3 | 8202 | Homo sapiens | Nuclear receptor coactivator 3 (NCOA3) |
| TARP | 445347 | Homo sapiens | TCR gamma alternate reading frame protein (TARP) |
| MDM2 | 4193 | Homo sapiens | MDM2 proto-oncogene (MDM2) |
| GHRL | 51738 | Homo sapiens | Ghrelin and obestatin prepropeptide (GHRL) |
| JMJD1C | 221037 | Homo sapiens | Jumonji domain containing 1C (JMJD1C) |
| CHFR | 55743 | Homo sapiens | Checkpoint with forkhead and ring finger domains (CHFR) |
| GSTP1 | 2950 | Homo sapiens | Glutathione S-transferase pi 1 (GSTP1) |
| DCC | 1630 | Homo sapiens | DCC netrin 1 receptor (DCC) |
| FKBP5 | 2289 | Homo sapiens | FK506 binding protein 5 (FKBP5) |
| MGMT | 4255 | Homo sapiens | O-6-methylguanine-DNA methyltransferase (MGMT) |
| CDH1 | 999 | Homo sapiens | Cadherin 1 (CDH1) |
| CDH3 | 1001 | Homo sapiens | Cadherin 3 (CDH3) |
| IGF1R | 3480 | Homo sapiens | Insulin like growth factor 1 receptor (IGF1R) |
| TNFRSF1A | 7132 | Homo sapiens | TNF receptor superfamily member 1A (TNFRSF1A) |
| MTHFR | 4524 | Homo sapiens | Methylenetetrahydrofolate reductase (MTHFR) |
| MDC1 | 9656 | Homo sapiens | Mediator of DNA damage checkpoint 1 (MDC1) |
| BCL2 | 596 | Homo sapiens | BCL2, apoptosis regulator (BCL2) |
| TGM2 | 7052 | Homo sapiens | Transglutaminase 2 (TGM2) |
| MTMR9 | 66036 | Homo sapiens | Myotubularin related protein 9 (MTMR9) |
| ERCC1 | 2067 | Homo sapiens | ERCC excision repair 1, endonuclease non-catalytic subunit (ERCC1) |
| APC | 324 | Homo sapiens | APC, WNT signaling pathway regulator (APC) |
| ERCC2 | 2068 | Homo sapiens | ERCC excision repair 2, TFIIH core complex helicase subunit (ERCC2) |
| FLT1 | 2321 | Homo sapiens | fms related tyrosine kinase 1 (FLT1) |
| GMDS | 2762 | Homo sapiens | GDP-mannose 4,6-dehydratase (GMDS) |
| VHL | 7428 | Homo sapiens | Von Hippel-Lindau tumor suppressor (VHL) |
| KLK3 | 354 | Homo sapiens | Kallikrein related peptidase 3 (KLK3) |
| TBX5 | 6910 | Homo sapiens | T-box 5 (TBX5) |
| IGF1 | 3479 | Homo sapiens | Insulin like growth factor 1 (IGF1) |
| IGF2 | 3481 | Homo sapiens | Insulin like growth factor 2 (IGF2) |
| BNC2 | 54796 | Homo sapiens | Basonuclin 2 (BNC2) |
| PERP | 64065 | Homo sapiens | PERP, TP53 apoptosis effector (PERP) |
GO enrichment analysis of EA
Functional enrichment analysis revealed a more specific function spectrum of these genes. Among the GO terms overrepresented in candidate genes were those associated with regulation and metabolic processes. In the BP category, terms directly associated with regulation and metabolic processes were identified, including organic substance metabolic process [Benjamini-Hochberg-corrected P (PBH) =1E-30], metabolic process (PBH =1E-30), cellular metabolic process (PBH =1E-30), and biological regulation (PBH =1E-30). Similarly, in the MF category, terms such as transcription factor binding (PBH =2.278E-08), receptor binding (PBH =9.10E-08), and protein binding (PBH =3.08E-13) were significantly enriched. In the CC category, the terms organelle lumen (PBH =8.73E-11), membrane-enclosed lumen (PBH =8.73E-11), and intracellular organelle lumen (PBH =8.73E-11) were extracted.
Enrichment of certain GO terms could be due to the contribution of other GO terms enriched at a lower hierarchy. Therefore, DAG was used to depict the results of GO enrichment analysis and how these affected other GO terms through upper hierarchies. For example, a part of the DAG representing BP (Figure 1A) was related with the GO term “biological regulation” in the rectangle. Surprisingly, this GO term was enriched at a very low FDR (<1e-20), and some other GO terms at lower hierarchies such as “regulation of biological process” and “regulation of cellular process” in the circle were enriched as a result. At the same, CC (Figure 1B) was associated with “intracellular”, and the lower terms such as “intracellular organelle” and “intracellular organelle part” were enriched. Moreover, in MF (Figure 1C), the GO term “regulatory region nucleic acid binding” was enriched and so were its upper terms “nucleic acid binding” and “organic cyclic compound binding”.
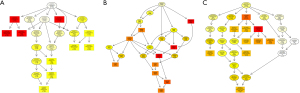
We used the ggplot2 package in Goseq software to depict the top 10 terms of BP, CC, and MF in graphs (Figure 2A-2C). For example, at the far left of the BP table, there were 4 biggest points representing “organic substance metabolic process”, “metabolic process”, “cellular metabolic process”, and “biological regulation” with the P value less than 1e-20 and count of EA genes more than 90. Interestingly, the most significant dot here was also the most red in DAG. The same results could be observed in CC and MF.

Pathway enrichment analysis of EA
Detection of the biological pathways enriched in the candidate genes may provide important information about the pathogenic molecular mechanism underlying EA. Through KEGG analysis, we found 177 significant enrichment pathways and extracted 19 most significant enrichment pathways (FDR ≤0.05) (Table 2). Among them, several pathways related to cancer, including bladder cancer (PBH =1.702E-6), prostate cancer (PBH =8.588E-5), pancreatic cancer (PBH =1.741E-4), chronic myeloid leukemia (PBH =1.079E-3), and small cell lung cancer (PBH =1.017E-2). In addition, drug reaction and metabolism-related processes were identified, such as platinum drug resistance (PBH =1.371E-4) and microRNAs in cancer (PBH =0.030). Further, pathways associated with cell growth and survival, including the p53 signaling pathway (PBH =0.003) and epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance (PBH =0.002), were also detected. The KEGG results in table 2 showed that “pathways in cancer” had an EA gene count of more than 30.
Table 2
| Pathway name | Database | ID | Input number | Background number | P value | Corrected P value | Gene ID | Gene name |
|---|---|---|---|---|---|---|---|---|
| Bladder cancer | KEGG PATHWAY | hsa05219 | 13 | 41 | 9.62E-09 | 1.70E-06 | 4312|4313|7157|999|1029|4193|1956|3845|1019|7422|4609|2064|1950 | MMP1|MMP2|TP53|CDH1|CDKN2A|MDM2|EGFR|KRAS|CDK4|VEGFA|MYC|ERBB2|EGF |
| Prostate cancer | KEGG PATHWAY | hsa05215 | 15 | 89 | 9.70E-07 | 8.59E-05 | 842|596|7157|3480|2064|3479|5290|4193|898|354|1956|3845|1950|2263|2950 | CASP9|BCL2|TP53|IGF1R|ERBB2|IGF1|PIK3CA|MDM2|CCNE1|KLK3|EGFR|KRAS|EGF|FGFR2|GSTP1 |
| Melanoma | KEGG PATHWAY | hsa05218 | 13 | 71 | 2.27E-06 | 0.000134 | 1021|7157|3480|1029|3479|5290|4193|999|1956|3845|1019|4233|1950 | CDK6|TP53|IGF1R|CDKN2A|IGF1|PIK3CA|MDM2|CDH1|EGFR|KRAS|CDK4|MET|EGF |
| Pathways in cancer | KEGG PATHWAY | hsa05200 | 33 | 397 | 3.02E-06 | 0.000134 | 596|4089|5290|5743|1630|25|7157|4233|3479|1019|324|2263|7474|4312|4313|898|3480|999|4193|1956|1950|4609|2950|1021|842|841|861|1029|354|3845|7422|2064|7428 | BCL2|SMAD4|PIK3CA|PTGS2|DCC|ABL1|TP53|MET|IGF1|CDK4|APC|FGFR2|WNT5A|MMP1|MMP2|CCNE1|IGF1R|CDH1|MDM2|EGFR|EGF|MYC|GSTP1|CDK6|CASP9|CASP8|RUNX1|CDKN2A|KLK3|KRAS|VEGFA|ERBB2|VHL |
| Platinum drug resistance | KEGG PATHWAY | hsa01524 | 13 | 75 | 3.87E-06 | 0.000137 | 842|841|596|7157|2064|1029|4193|5290|2947|2944|2950|2952|2067 | CASP9|CASP8|BCL2|TP53|ERBB2|CDKN2A|MDM2|PIK3CA|GSTM3|GSTM1|GSTP1|GSTT1|ERCC1 |
| Pancreatic cancer | KEGG PATHWAY | hsa05212 | 12 | 66 | 5.90E-06 | 0.000174 | 1021|842|7157|4089|1029|7422|5290|1956|3845|1019|2064|1950 | CDK6|CASP9|TP53|SMAD4|CDKN2A|VEGFA|PIK3CA|EGFR|KRAS|CDK4|ERBB2|EGF |
| Non-small cell lung cancer | KEGG PATHWAY | hsa05223 | 11 | 56 | 7.55E-06 | 0.000191 | 1021|842|2272|7157|1029|5290|1956|3845|1019|2064|1950 | CDK6|CASP9|FHIT|TP53|CDKN2A|PIK3CA|EGFR|KRAS|CDK4|ERBB2|EGF |
| Endometrial cancer | KEGG PATHWAY | hsa05213 | 10 | 52 | 2.30E-05 | 0.000508 | 842|7157|999|5290|1956|3845|1950|324|4609|2064 | CASP9|TP53|CDH1|PIK3CA|EGFR|KRAS|EGF|APC|MYC|ERBB2 |
| Glioma | KEGG PATHWAY | hsa05214 | 11 | 65 | 2.60E-05 | 0.000511 | 1021|7157|3480|1029|3479|5290|4193|1956|3845|1019|1950 | CDK6|TP53|IGF1R|CDKN2A|IGF1|PIK3CA|MDM2|EGFR|KRAS|CDK4|EGF |
| Endocrine resistance | KEGG PATHWAY | hsa01522 | 13 | 97 | 4.52E-05 | 0.000801 | 4313|8202|596|7157|3480|1029|3479|5290|4193|1956|3845|1019|2064 | MMP2|NCOA3|BCL2|TP53|IGF1R|CDKN2A|IGF1|PIK3CA|MDM2|EGFR|KRAS|CDK4|ERBB2 |
| Chronic myeloid leukemia | KEGG PATHWAY | hsa05220 | 11 | 73 | 6.71E-05 | 0.001079 | 25|1021|7157|4089|1029|4193|5290|861|3845|1019|4609 | ABL1|CDK6|TP53|SMAD4|CDKN2A|MDM2|PIK3CA|RUNX1|KRAS|CDK4|MYC |
| EGFR tyrosine kinase inhibitor resistance | KEGG PATHWAY | hsa01521 | 11 | 81 | 0.000154 | 0.002278 | 596|3480|3479|5290|2064|1956|3845|1950|2263|4233|7422 | BCL2|IGF1R|IGF1|PIK3CA|ERBB2|EGFR|KRAS|EGF|FGFR2|MET|VEGFA |
| p53 signaling pathway | KEGG PATHWAY | hsa04115 | 10 | 69 | 0.00019 | 0.002585 | 1021|842|841|7157|1029|3479|4193|64065|898|1019 | CDK6|CASP9|CASP8|TP53|CDKN2A|IGF1|MDM2|PERP|CCNE1|CDK4 |
| HIF-1 signaling pathway | KEGG PATHWAY | hsa04066 | 12 | 103 | 0.000289 | 0.003654 | 2321|596|3480|7422|3479|5290|2064|1956|1950|4846|3162|7428 | FLT1|BCL2|IGF1R|VEGFA|IGF1|PIK3CA|ERBB2|EGFR|EGF|NOS3|HMOX1|VHL |
| Colorectal cancer | KEGG PATHWAY | hsa05210 | 9 | 62 | 0.00039 | 0.004601 | 1630|842|596|7157|4089|5290|3845|324|4609 | DCC|CASP9|BCL2|TP53|SMAD4|PIK3CA|KRAS|APC|MYC |
| Small cell lung cancer | KEGG PATHWAY | hsa05222 | 10 | 86 | 0.00092 | 0.010174 | 1021|842|2272|7157|596|5290|898|5743|1019|4609 | CDK6|CASP9|FHIT|TP53|BCL2|PIK3CA|CCNE1|PTGS2|CDK4|MYC |
| Central carbon metabolism in cancer | KEGG PATHWAY | hsa05230 | 8 | 67 | 0.002516 | 0.0262 | 7157|2064|5290|1956|3845|4609|2263|4233 | TP53|ERBB2|PIK3CA|EGFR|KRAS|MYC|FGFR2|MET |
| MicroRNAs in cancer | KEGG PATHWAY | hsa05206 | 20 | 299 | 0.003053 | 0.030023 | 25|4170|596|7157|3162|2064|7078|1029|4193|5290|5743|898|1956|3845|7422|324|4609|1021|4233|27086 | ABL1|MCL1|BCL2|TP53|HMOX1|ERBB2|TIMP3|CDKN2A|MDM2|PIK3CA|PTGS2|CCNE1|EGFR|KRAS|VEGFA|APC|MYC|CDK6|MET|FOXP1 |
| Proteoglycans in cancer | KEGG PATHWAY | hsa05205 | 15 | 205 | 0.004502 | 0.041943 | 4313|7078|7157|3480|2064|4193|5290|3479|3481|1956|3845|7422|4609|7474|4233 | MMP2|TIMP3|TP53|IGF1R|ERBB2|MDM2|PIK3CA|IGF1|IGF2|EGFR|KRAS|VEGFA|MYC|WNT5A|MET |
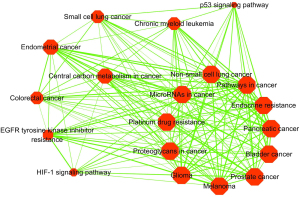
Crosstalk among significantly enriched pathways
To perform further analysis of the pathways and how they interact with each other, we implemented a pathway crosstalk analysis for the 19 enriched pathways. The screening conditions included more than 5 candidate genes per pathway and at least 3 genes for each pathway shared with 1 or more other pathways. Coincidentally, the 19 pathways met our screening criteria for crosstalk analysis. All of the 161 pathway pairs (edges) among the 19 pathways, which were ranked according to the average JC and OC scores, were used to construct the pathway crosstalk network. Based on the crosstalk, these pathways could be probably divided into 2 major modules, each of which contained pathways that had more crosstalk with each other compared with other pathways and were likely related to the similar biological processes or etiology (Figure 3). The first module included a variety of cancers, metabolism-related processes, and drug reaction pathways, such as bladder cancer, prostate cancer, pancreatic cancer, non-small cell lung cancer, endocrine resistance, microRNAs in cancer, proteoglycans in cancer, and platinum drug resistance. In the other module, cell transcriptional regulation predominated, including endometrial cancer, chronic myeloid leukemia, p53 signaling, HIF-1 signaling, and central carbon metabolism in cancer. As the above results indicated, pathway crosstalk analysis could provide important insight into the understanding of oncogenic mechanisms.
EA-specific protein network
To distill insight into the potential pathological protein network of EA, we constructed a subnetwork for EA from the human PPI network with the Klein-Ravi algorithm. Typically, this algorithm will connect as many input nodes as possible (genes included in EA genes) with the minimum number of interlinking nodes. As shown in Figure 4, the protein network of EA contained 116 nodes and 284 edges. Of the 109 EA genes, 104 were included in the EA-specific network, which accounted for about 95.4% of the EA genes and 89.7% of the genes in the EA-specific network, indicating a high coverage of EA genes in the subnetwork. In addition to EA genes, a further 12 genes were contained in the EA-specific network (Table 3). In view of the close interaction of these genes with reported EA-related genes, they may also participate in the pathological process of EA. Notably, many of the EA-related genes, including TYMS, GSTO1, and BMP7, have been reported in previous studies (10-12). While some of these genes have not been shown to directly participate in the pathophysiology of EA, some of the genes linked to them or other member genotypes of the same protein family have been found to be involved in these reactions.
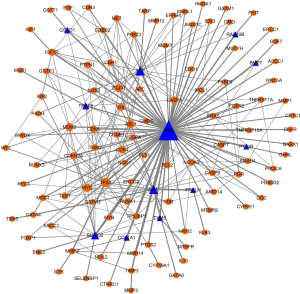
Table 3
| Gene abbreviations | Gene ID | Species | Gene name |
|---|---|---|---|
| PTHLH | 5744 | Homo sapiens | parathyroid hormone like hormone (PTHLH) |
| SUMO2 | 6613 | Homo sapiens | small ubiquitin-like modifier 2 (SUMO2) |
| TYMS | 7298 | Homo sapiens | thymidylate synthetase (TYMS) |
| APP | 351 | Homo sapiens | amyloid beta precursor protein (APP) |
| PTGIR | 5739 | Homo sapiens | prostaglandin I2 (prostacyclin) receptor (IP) (PTGIR) |
| SP1 | 6667 | Homo sapiens | Sp1 transcription factor (SP1) |
| UBC | 7316 | Homo sapiens | ubiquitin C (UBC) |
| COL1A1 | 1277 | Homo sapiens | collagen type I alpha 1 chain (COL1A1) |
| GSTO1 | 9446 | Homo sapiens | glutathione S-transferase omega 1 (GSTO1) |
| TRAF6 | 7189 | Homo sapiens | TNF receptor associated factor 6 (TRAF6) |
| BMP7 | 655 | Homo sapiens | bone morphogenetic protein 7 (BMP7) |
| RAB40B | 10966 | Homo sapiens | RAB40B, member RAS oncogene family (RAB40B) |
Discussion
Recently, considerable progress has been made in the study of the molecular mechanisms of EA. Advances in technology have allowed us to identify more and more genes and proteins associated with this disease on a larger scale. While a large number of studies have reported on the pathogenesis genes of EA, especially in the progression of Barrett’s esophagus to EA, in-depth analysis of the biochemical processes related to the pathogenesis of EA at the molecular level is still limited. Besides, there is no literature to summarize and analyze the relationship between these genes and EA. As a result, a systematic analysis of EA-related genes based on pathways and networks would provide a valuable resource for analyzing gene function, biochemical pathways, and subnetworks of EA (13). In this study, we pooled and managed data from a number of studies on human genes associated with EA to provide a comprehensive analysis of EA-related biochemical processes and their interrelations.
Functional enrichment analysis has brought the specific biological processes involved in EA genes to light. Our results showed that genes involved in EA might play an important role in metabolic processes, cell growth and survival, and drug reactions. The terms cellular metabolic process, organic substance metabolic process, biological regulation, regulation of cell death, regulation of apoptotic process, and response to stimulus were significantly enriched in EA genes, suggesting they play an important role in the pathogenesis of EA.
Pathway analysis showed that various cancer pathways were enriched in EA genes, such as bladder cancer, prostate cancer, pancreatic cancer, and so forth. This might be due to the regulation of the same genes among various cancers and also confirmed that the occurrence of cancer is not a single molecular event but a multistep process. Meanwhile, it was noteworthy that almost all of these cancer pathways contained TP53 and PIK3CA. As a well-known tumor suppressor gene, TP53, also called p53, can induce cell cycle arrest to repair DNA damage or lead to apoptosis when growth arrest is not realized. However, after p53 deletion or when mutated cells undergo DNA damage, the cells continue to increase, and hence the abnormality of DNA is transmitted to the progeny cells. More than 50% of human tumors carry mutations in the p53 gene (14). PIK3CA encodes the p110α catalytic subunit of the class IA phosphatidylinositol 3-kinases (PI3Ks) and then initiates a complex signaling cascade reaction that can result in cell proliferation, survival, and regulation of motility, among others (15). At the same time, some pathways were associated with cell growth and survival. For example, as a transcription factor, HIF-1 plays an important role in the formation of blood vessels and the proliferation of tumor cells (16). The generation of tumor blood vessels brings oxygen and nutrients to tumor cells, promotes the growth and proliferation of tumor cells, and is the basis for tumor cell invasion and metastasis. In addition, EGFR tyrosine kinase inhibitors block the abnormal signal transduction of EGFR and tyrosine kinase and induce apoptosis, thereby inhibiting the growth of tumor cells. EGFR tyrosine kinase inhibitors produce resistance via mechanisms such as EGFR mutations (17). Further, proteoglycans, as a sort of extracellular matrix in the tumor microenvironment, regulate the biological behavior of tumor cells via metabolic processes (18). These results suggested that the molecular mechanism of EA was rather intricate, involving many genes, pathways, and their interactions.
Importantly, in pathway crosstalk analysis, we established 2 major modules, of which 1 was mainly related to metabolic regulation and drug reactions. Among the pathways, we noted that many were tumor-associated pathways involved in metabolism. For example, the MMP1 and MMP2 genes in bladder cancer are members of the matrix metalloproteinase (MMP) gene family, which are involved in the breakdown of the extracellular matrix and play an important role in physiological processes, and thus their dysfunction is related to many diseases (including tumors) (19). In addition, the protein encoded by insulin like growth factor 1 (IGF1), which is found in prostate cancer, melanoma, and glioma, is similar to insulin in function and structure but has much higher growth promoting activity and is related to tumors (20). Similarly, platinum drug resistance, microRNAs, and endocrine resistance involved in metabolic regulation and drug reactions have been well studied (21). MicroRNAs directly or indirectly regulate several key enzymes and/or signaling hubs that result in the altering of metabolic pathways, leading to tumor progression and/or metastasis (22). Further, we noted that the 2 pathway modules were related to each other, and the genes in the pathways also interacted, suggesting that there was a transcription-metabolic regulatory network in the molecular mechanism of EA.
In addition, we extracted EA-specific protein networks based on the human PPI network. It was worth noting that some genes did not belong to EA genes but were contained in the human PPI network which may be related to EA. For instance, RAB40B may be involved in tissue-supporting basement membrane or extracellular matrix destruction, and its encoded protein may regulate secretory vesicles, which may participate in tumor metastasis (23). NF-κB is activated in cells that become malignant tumors and cells that are recruited to and constitute the tumor microenvironment. TRAF6 can serve as its activation molecule, leading to the expression of abnormal genes and malignancy (24). TYMS has recently been spotlighted as a target of cancer chemotherapeutics, particularly 5-FU, considering that it plays a role in DNA replication and repair (25). SUMO2 plays a crucial role in nuclear transport, DNA replication and repair, mitosis, and signal transduction (26). As the results showed, network-based analysis could provide subnetworks for EA genes and also have the potential to detect promising related genes.
However, our study had some limitations. First, our analysis was based on reported EA-related genes from existing literature and as a result, the potential biases and deficiencies in the available reports inevitably affected our result. At the same time, there were few studies on the molecular mechanism of EA, and consequently, we might have omitted a number of important processes related to EA. Although the overall quality of PPI databases has been greatly improved, current technical limitations remain, and PPI data may also contain some false-positive results (27). With PPI data becoming more comprehensive and accurate, an EA-specific subnetwork will become more valuable.
Conclusions
We performed a comprehensive investigation into the pathways and molecular network of EA based on the EA genes. Integrating information from biological functions, biochemical pathways, and pathway crosstalk analyses, we found that biochemical processes and pathways associated with metabolic processes, biological regulation processes, and the signal transduction process played key roles in the molecular mechanism of EA. In addition, there were 2 interconnected pathway modules: 1 which included metabolic regulation and drug reactions, and the other transcriptional regulation. Furthermore, we extracted an EA-specific subnetwork and predicted some novel genes potentially bound up with EA. Our results provided critical information for further analysis and indicated that system level analysis was promising for exploring the molecular mechanisms of EA.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1286/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1286/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lin Y, Wang HL, Fang K, et al. International trends in esophageal cancer incidence rates by histological subtype (1990-2012) and prediction of the rates to 2030. Esophagus 2022;19:560-8. [Crossref] [PubMed]
- Coleman HG, Xie SH, Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology 2018;154:390-405. [Crossref] [PubMed]
- Hoppe S, Jonas C, Wenzel MC, et al. Genomic and Transcriptomic Characteristics of Esophageal Adenocarcinoma. Cancers (Basel) 2021;13:4300. [Crossref] [PubMed]
- Noordman BJ, van Klaveren D, van Berge Henegouwen MI, et al. Impact of Surgical Approach on Long-term Survival in Esophageal Adenocarcinoma Patients With or Without Neoadjuvant Chemoradiotherapy. Ann Surg 2018;267:892-7. [Crossref] [PubMed]
- Rice ES, Green RE. New Approaches for Genome Assembly and Scaffolding. Annu Rev Anim Biosci 2019;7:17-40. [Crossref] [PubMed]
- Qi W, Li R, Li L, et al. Identification of key genes associated with esophageal adenocarcinoma based on bioinformatics analysis. Ann Transl Med 2021;9:1711. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Cowley MJ, Pinese M, Kassahn KS, et al. PINA v2.0: mining interactome modules. Nucleic Acids Res 2012;40:D862-5. [Crossref] [PubMed]
- Zheng S, Zhao Z. GenRev: exploring functional relevance of genes in molecular networks. Genomics 2012;99:183-8. [Crossref] [PubMed]
- Peng DF, Razvi M, Chen H, et al. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett's adenocarcinoma. Gut 2009;58:5-15. [Crossref] [PubMed]
- Langer R, Specht K, Becker K, et al. Comparison of pretherapeutic and posttherapeutic expression levels of chemotherapy-associated genes in adenocarcinomas of the esophagus treated by 5-fluorouracil- and cisplatin-based neoadjuvant chemotherapy. Am J Clin Pathol 2007;128:191-7. [Crossref] [PubMed]
- Rees JR, Onwuegbusi BA, Save VE, et al. In vivo and in vitro evidence for transforming growth factor-beta1-mediated epithelial to mesenchymal transition in esophageal adenocarcinoma. Cancer Res 2006;66:9583-90. [Crossref] [PubMed]
- Rezaei-Tavirani M, Rezaei-Taviran S, Mansouri M, et al. Protein-Protein Interaction Network Analysis for a Biomarker Panel Related to Human Esophageal Adenocarcinoma. Asian Pac J Cancer Prev 2017;18:3357-63. [Crossref] [PubMed]
- Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat 2014;35:672-88. [Crossref] [PubMed]
- Essakly A, Loeser H, Kraemer M, et al. PIK3CA and KRAS Amplification in Esophageal Adenocarcinoma and their Impact on the Inflammatory Tumor Microenvironment and Prognosis. Transl Oncol 2020;13:157-64. [Crossref] [PubMed]
- Igbo BT, Linge A, Frosch S, et al. Immunohistochemical analyses of paraffin-embedded sections after primary surgery or trimodality treatment in esophageal carcinoma. Clin Transl Radiat Oncol 2022;36:106-12. [Crossref] [PubMed]
- Izadi F, Sharpe BP, Breininger SP, et al. Genomic Analysis of Response to Neoadjuvant Chemotherapy in Esophageal Adenocarcinoma. Cancers (Basel) 2021;13:3394. [Crossref] [PubMed]
- Theocharis AD, Skandalis SS, Tzanakakis GN, et al. Proteoglycans in health and disease: novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J 2010;277:3904-23. [Crossref] [PubMed]
- Decock J, Paridaens R, Ye S. Genetic polymorphisms of matrix metalloproteinases in lung, breast and colorectal cancer. Clin Genet 2008;73:197-211. [Crossref] [PubMed]
- Dighe SG, Chen J, Yan L, et al. Germline variation in the insulin-like growth factor pathway and risk of Barrett's esophagus and esophageal adenocarcinoma. Carcinogenesis 2021;42:369-77. [Crossref] [PubMed]
- Gambardella V, Fleitas T, Tarazona N, et al. Towards precision oncology for HER2 blockade in gastroesophageal adenocarcinoma. Ann Oncol 2019;30:1254-64. [Crossref] [PubMed]
- Petrick JL, Pfeiffer RM, Liao LM, et al. Circulating MicroRNAs in Relation to Esophageal Adenocarcinoma Diagnosis and Survival. Dig Dis Sci 2021;66:3831-41. [Crossref] [PubMed]
- Li Y, Jia Q, Wang Y, et al. Rab40b upregulation correlates with the prognosis of gastric cancer by promoting migration, invasion, and metastasis. Med Oncol 2015;32:126. [Crossref] [PubMed]
- Wang J, Wu X, Jiang M, et al. Mechanism by which TRAF6 Participates in the Immune Regulation of Autoimmune Diseases and Cancer. Biomed Res Int 2020;2020:4607197. [Crossref] [PubMed]
- Varghese V, Magnani L, Harada-Shoji N, et al. FOXM1 modulates 5-FU resistance in colorectal cancer through regulating TYMS expression. Sci Rep 2019;9:1505. [Crossref] [PubMed]
- Bursomanno S, Beli P, Khan AM, et al. Proteome-wide analysis of SUMO2 targets in response to pathological DNA replication stress in human cells. DNA Repair (Amst) 2015;25:84-96. [Crossref] [PubMed]
- Zhang Z, Jiang M, Wu D, et al. A Novel Method for Identifying Essential Proteins Based on Non-negative Matrix Tri-Factorization. Front Genet 2021;12:709660. [Crossref] [PubMed]
(English Language Editor: A. Muylwyk)


