
Whole-genome sequencing of 20 cholangiocarcinoma cases reveals unique profiles in patients with cirrhosis and primary sclerosing cholangitis
Highlight box
Key findings
• SBS17 is significantly enriched in cases of cholangiocarcinoma that arise in patients with underlying ulcerative colitis and primary sclerosing cholangitis.
• TP53 mutations are significantly enriched in cholangiocarcinoma that arise in an inflamed liver and significantly correlate with short term survival.
• The mutational profile of short-term survivors is similar to extrahepatic cholangiocarcinoma.
What is known and what is new?
• Cholangiocarcinoma is a molecularly heterogenous disease with unique molecular alterations across histological and anatomical subtypes.
• We describe mutational alterations unique to short term survivors and cholangiocarcinomas that arise in patients with inflammatory liver disease.
What is the implication, and what should change now?
• WGS provides additional biological information regarding the heterogenous nature of CCA.
• Ongoing observational studies are required to describe unique genetic signatures and molecular alterations seen in cholangiocarcinoma arising in patients with underlying inflammatory disorders.
Introduction
Cholangiocarcinoma (CCA) is a lethal malignancy originating from epithelial cells lining the biliary tree (1). CCA is divided anatomically into intrahepatic (iCCA), perihilar (pCCA), and distal cholangiocarcinoma (dCCA). Morphologically, iCCA is further divided into mass-forming, periductal infiltrating, and intraductal growth types (2). Within iCCA, histopathological subdivision also identifies small duct (SD) and large duct (LD) types. Observational studies have revealed different clinical outcomes amongst the different subtypes of CCA and recent studies describing the mutational landscape has aimed to explain these differences and better classify CCA at the molecular level. Patients with SD-iCCA are known to have favourable outcomes compared to those with LD histology. They have higher response rates to conventional chemotherapy and improved overall survival (OS) (3). Genetic analysis has revealed the presence of IDH1/2 mutations and FGFR fusions in SD-iCCA. At the molecular level, LD-iCCA lack IDH1/2 and FGFR fusions and are more similar to pCCA and dCCA and have frequent KRAS, SMAD4, CDKN2A, and TP53 mutations (4-6).
Resection is the major curative approach for CCA and the recent BILCAP (capecitabine compared with observation in resected biliary tract cancer) trial has emphasized the benefits of adjuvant capecitabine (7). In select patients with iCCA or pCCA, liver transplantation can also be curative (8). In advanced stage disease, systemic treatment options typically involve a combination of cisplatin and gemcitabine (9) and/or taxane-based palliative chemotherapy (10) with a median survival time less than 1 year. Despite this, molecular profiling studies in CCA have identified targetable alterations including IDH1 mutations and FGFR2 fusions which are enriched in iCCA and are likely both prognostic and predictive (11).
There are various risk factors for iCCA yet associations between molecular profiles and aetiology remains largely undetermined. Although CCA often develops in a non-cirrhotic liver (12), chronic liver disease and chronic inflammatory states are well known risk factors. These include fluke infections, cirrhosis and primary sclerosing cholangitis (PSC) (13,14). Genomic characterisation of CCA has highlighted a high prevalence of age associated mutational signatures and enrichment of APOBEC mutagenesis particularly in Fluke positive CCA (15).
It has been well documented that patients with PSC and CCA have a particularly poor prognosis and most patients die within one year of diagnosis (16). Limited studies suggest that CCA arising in an inflammatory/autoimmune setting may closely resemble dCCA at a molecular level and harbour genomic alterations associated with poor prognosis such as TP53 mutations and HER2 amplifications, while lacking alterations associated with more indolent disease such as FGFR2 translocations (17,18). In this study, we sought to perform whole genome sequencing (WGS) in patients with short-term survival (STS), enriching for patients with cirrhosis and patients with PSC, and we compared the molecular profiles to patients with longer-term survival (LTS). We present the following article in accordance with the STROBE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-676/rc).
Methods
Sample collection
A Biliary Tract Cancer (BTC) database at the Princess Margaret Cancer Centre (PMCC) was utilized to identify patients with underlying cirrhosis or PSC (19). Considering the BILCAP trial findings, additional patients with resected CCA and very short survival were included (survival <18 months post-surgical resection). Patients with long survival were used as a comparator (survival >51 months). Informed consent was previously obtained at the time of surgical resection according to institutional review board approved protocols. Once patients were identified, fresh-frozen tissue was obtained from the UHN biobank. Samples were reviewed by a pathologist who confirmed the histological diagnosis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Sample processing and sequencing
All samples underwent tumour enrichment using laser capture microdissection (LCM) as previously described (20,21). DNA extraction was performed at the UHN biobank laboratory. Library preparation, sequencing, and bioinformatic analysis was performed at the Ontario Institute for Cancer Research (OICR) (20,21).
WGS was performed as described elsewhere (20-22). DNA was quantified using Qubit dsDNA kit per the manufacturers protocol. The NEB Next DNA sample preparation kit was used to generate paired-end libraries. Cluster generation and sequencing was performed using the Illumina HiSeq 2000/2500 platform with TruSeq Cluster kit v3 (Illumina Inc., San Diego, CA, USA Cat #PE-401-3001/FC-401-3001).
The Burrow-Wheeler Aligner (BWA, version 0.7.17) was used to align the raw sequencing reads to the human reference genome build hg38. The Genome Analysis Toolkit (GATK4, version 4.1.2) was used to detect germline mutations (23). Somatic single nucleotide variations (SNVs) and Indels were identified using Strelka2 (version 2.9.10) (24), MuTect2 (version 4.1.2) (25), SVaBA (v134), and DELLY2 (version 0.8.1). Copy number segments and tumour cellularity were obtained using a custom algorithm, “Celluloid” (20). Structural rearrangements were identified using Manta (version 1.6.0), SVaBA (v134), and DELLY2 (version 0.8.1).
Mutational signatures were identified using a non-negative matrix factorization method of base substitution (21,26). The contribution and the significance of these mutational signatures in each sample was determined by applying a non-negative least squares linear model, using the published signatures as independent variables. Significance was assessed using 5,000 bootstrap replicates of the mutation counts.
HRDetect scores were applied to samples as previously published, with a score of >0.7 predicting homologous recombination deficiency (HRD) (27).
Statistical analyses
Descriptive analyses were used to report clinical and pathological characteristics. Compiled data were analysed using a two-tailed t-test or Fisher’s exact test where indicated. OS was calculated from the time of resection or first pathological diagnosis until date of death or last follow-up. For the t-test, P values <0.05 were considered statistically significant.
Results
WGS was performed on 20 pathologically confirmed CCAs and matched whole blood samples (Table 1). These included 14 patients with iCCA, and 6 patients with pCCA. Of the 20 samples, 7 were from patients with underlying cirrhosis or PSC (STS group) including 4 cases with PSC, 2 with hepatitis B cirrhosis, and 1 with hemochromatosis-cirrhosis (Table 1). An additional 3 resected CCA with no known liver disease but short survival were included in this group (Table 1). Median tumour cellularity was 83.5% (range, 36.3–97%) (Table S1) and median sequencing depth of tumour samples was 46.8X (range, 30.2X to 56.5X) and 32.9X (range, 29.1X to 38.9X) in matched whole blood samples. The median OS in the STS group was 13 months (95% CI: 2–17 months). In the LTS group, the median follow-up was 123 months and median survival had not yet been reached with 8 of 10 patients alive and disease-free after 8 years. Most LTS and STS samples were early stage CCA. The LTS group included 9 patients with stage I or stage II disease and 1 patient with stage IIIA disease (T3N0, AJCC v8) while the STS group included 6 patients with stage II disease and 2 patients with stage III disease (T2N1 AJCC v8). Two additional patients with locally advanced unresectable disease were included. BTC_8002 was a 26-year-old male with T2N0 (stage II) disease while PANX_1237 was a 45-year-old male with T3N0 (stage IIIA) disease. More patients in the STS group had poorly differentiated tumours (n=5/10) compared to LTS (n=1/10). This association was non-significant (P=0.14).
Table 1
| Samples | Tumour Type | Age at diagnosis, years | OS [months] | Sex | Chronic liver disease/STS | Stage/differentiation | Driver alterations | TMB | Dominant single base substitution signatures |
|---|---|---|---|---|---|---|---|---|---|
| BTC_8002 | iCCA | 26 | 3 | M | STS: UC/PSC | LA/poor | TP53 R282fs; SMAD4 c.425-2A>G | 12.61 | 28 (55%), 17 (25%) |
| BTC_9001 | iCCA | 40 | 16 | F | STS: UC/PSC | II/poor | TP53 R248Q, CDKN2A Del | 2.95 | 1 (36%), 8 (20%), 9 (18%), 17 (7%) |
| BTC_9004 | iCCA | 51 | 16 | F | STS: HepB cirrhosis | II/poor | TP53 E11fs; RET T75M; ERBB2 amplification; AKT2 Amplification | 2.71 | 8 (24%), 12 (21%), 9 (14%), 19 (13%) |
| BTC_9005 | iCCA | 67 | 16 | M | STS: HepB cirrhosis | II/moderate | TP53 R175L, CDKN2A Del, BAP1 I589fs + LOH | 1.80 | 5 (54%), 1 (16%), 8 (14%) |
| BTC_9009 | iCCA | 72 | 10 | F | STS: PSC | II/well | KRAS G12R; TP53 Y220C; MYC Amp | 2.14 | 1 (46%), 8 (30%), 9 (16%) |
| BTC_9011 | iCCA | 78 | 10 | F | STS | IIIB/poor | TP53 R175H; TGFBR2 T409fs; ERBB3 A42T; ARID1A P145fs + LOH |
33.23 | 1 (47%), 6 (21%), 21 (13%) |
| BTC_9016 | pCCA | 58 | 15 | M | STS | IIIC/well | CDKN2A Del; PIK3CA E726K; MDM2 Amp | 0.87 | 1 (40%), 16 (27%) 8 (24%) |
| BTC_9017 | iCCA | 66 | 10 | M | STS: hemochromatosis- cirrhosis | II/moderate | TP53 R273C; ARID1A Q1974X + LOH; TGFBR2 C102S | 1.85 | 5 (53%), 8 (28%) |
| BTC_9018 | pCCA | 83 | 17 | M | STS | II/moderate | BRAF G469A; SMAD4 L535fs; ERBB3 V104L; cKIT Y543H | 7.06 | 16 (43%), 9 (23%), 1 (17%), 3 (13%) |
| PANX_1237 | iCCA | 45 | 2 | M | STS: UC/PSC | LA/poor | RAF-TRIM fusion; TP53 Q192X; CDKN2A E88X; SMAD4 I74fs | 4.02 | 9 (22%), 8 (20%), 2 (15%), 13 (15%), 1 (14%), 17 (5%) |
| BTC_9002 | iCCA | 72 | Alive [152] | F | LTS | II/well | FGFR2 Fusion | 1.18 | 5 (35%), 16 (32%), 1 (20%), 8 (12%) |
| BTC_9003 | iCCA | 57 | Alive [132] | M | LTS | II/moderate | PBRM1 Y81X + LOH; BAP1 V27fs + LOH | 1.37 | 5 (51%), 1 (15%), 18 (10%) |
| BTC_9006 | pCCA | 57 | Alive [102] | F | LTS | I/well | KRAS Q61H, CDKN2A deletionSV; CTNNB1 D32G | 1.53 | 5 (40%), 8 (22%),1 (14%), 9 (11%) |
| BTC_9007 | iCCA | 62 | Alive [108] | F | LTS | I/well | BRAF V600E | 0.73 | 5 (55%), 4 (20%), 8 (13%), 1 (10%) |
| BTC_9010 | pCCA | 52 | Alive [127] | F | LTS | II/moderate | PBRM1 SV + LOH; BAP1 57_64del | 1.29 | 5 (53%), 8 (19%), 1 (14%) |
| BTC_9012 | iCCA | 57 | Alive [172] | M | LTS | I/well | FGFR2 Fusion | 1.03 | 8 (23%), 5 (22%), 30 (21%), 16 (20%), 1 (13%) |
| BTC_9013 | pCCA | 50 | Alive [162] | F | LTS | I/moderate | PIK3CA E365K; BAP1 T164fs + LOH | 1.33 | 1 (34%), 16 (28%), 8 (20%) |
| BTC_9014 | iCCA | 64 | 80 | M | LTS | II/moderate | NRAS Q61R | 1.15 | 5 (48%), 8 (20%), 1 (19%) |
| BTC_9015 | pCCA | 54 | Alive [120] | M | LTS | IIIA/well | BAP1 R150fs + LOH | 1.69 | 16 (40%), 1 (26%), 8 (12%), 30 (10%) |
| BTC_9019 | iCCA | 32 | 55 | F | LTS | II/poor | 2.06 | 5 (46%), 8 (23%), 4 (12%) |
BTC, biliary tract cancer; iCCA, intrahepatic cholangiocarcinoma; pCCA, perihilar cholangiocarcinoma; OS, overall survival; M, male; F, female; STS, short term survivor; UC, ulcerative colitis; PSC, primary sclerosing cholangitis; HepB, hepatitis B; LA, locally advanced; LTS, long term survivor; LOH, loss of heterozygosity; TMB, tumour mutation burden.
Driver mutations and copy number alterations
Given the enriched population of patients with underlying chronic inflammatory disease and cirrhosis, we observed trends in recurrent driver alterations in the STS group. TP53 inactivating mutations were significantly enriched for in the STS population (P<0.01, 2-sided Fisher’s exact test). WGS identified inactivating TP53 mutations in 8/10 (80%) STS samples and in no samples from the LTS group (Table 1, Figure 1). Notably, all patients (n=7) with PSC/Hep-B cirrhosis and haemachromatosis-cirrhosis had TP53 inactivating mutations (Table 1). Other recurrent alterations in the STS group included loss of CDKN2A (n=5/10) and SMAD4 (n=3/10). In comparison, alterations in the LTS group included BAP1 (n=4), BRAF V600E, predicted FGFR fusions (n=2), and an IDH1 mutation (n=1).

Alterations in the chromatin remodelling genes ARID1A, PBRM1, and BAP1 were found in 7/20 (35%) samples (Table 1). Other potentially targetable alterations included PIK3CA non-synonymous point mutations, ERBB3 p.A42T, RAF-TRIM fusion, and a cKIT Y543H mutation. One patient with known Lynch Syndrome was included in the STS cohort and harboured a germline pathogenic variant in MLH1 (G67R). Focal copy number amplifications in MYC (n=1), MDM2 (n=1), and AKT2 (n=1) were also identified through WGS and found exclusively in the STS group.
Mutational signatures and clinical correlations
WGS was used to detect the presence of single base substitution (SBS) cosmic signatures across tumour samples. Overall, the samples were dominated by age associated signatures SBS1 and SBS5 (Table 1). Notably, mutations attributed to SBS5 were more common in LTS compared to STS (Table 1). In our selected dataset, SBS5 did not significantly correlate with patient age at diagnosis (r=−0.19). SBS17, a signature of unknown aetiology, was documented in 3/20 patients (Table 1). All three of these patients had underlying ulcerative colitis (UC) with PSC and SBS17 significantly correlated with PSC status (P<0.01, 2-sided Fishers exact test). Patients with SBS17 were much younger than the rest of our cohort with a median age of 40 vs. 58 years (P=0.006).
The median tumour mutation burden (TMB) across the cohort was 1.75 mutations/Mb (range, 0.73–33.23). The patient (BTC_9011) with known Lynch Syndrome (MLH1 p.G67R) included in the STS group had a significantly elevated TMB of 33 mut/Mb (Figure 2A). Notably this case harboured an inactivating point mutation in TP53 (p.R175H) and a frameshift mutation in JAK2. SBS6 and SBS26, two signatures associated with defects in DNA mismatch repair (MMR) were exclusively found in this patient (Table 1). This patient had previously been diagnosed with colon cancer treated with colectomy and endometrial cancer treated with total abdominal hysterectomy (TAH) with bilateral salpingo-oophorectomy (BSO). She declined any systemic therapy.
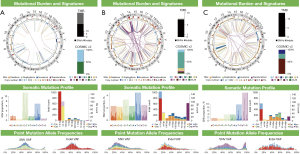
BTC_9018 (STS group), exhibited numerous genomic features suggestive of HRD including a high SNV load of 21,158, a high structural variant load of 259 and a large number of 4bp+ deletions (Figure 2B). This patient had a prior history of mantle cell lymphoma treated with rituximab, cyclophosphamide, vincristine, and prednisone (R-CVP), having completed two years of maintenance rituximab just prior to diagnosis. SBS3, the characteristic signature of HRD was detected in this case (13% of mutations) (Table 1). Notably, this sample did not harbour any germline or somatic alterations in DNA damage response genes (Table S2). We calculated HRDetect scores to further determine the likelihood of HRD in this sample and throughout our cohort (Table S3). Of the 20 samples in our cohort, this case had the highest score (intermediate score of 0.47) but did not meet the cut-off of 0.7 generally considered to define HRD cases. The patient did not receive systemic therapy due to performance status.
PSC-CCA
Four cases of PSC with iCCA were profiled as part of the STS group. Interestingly, SBS17 was clearly evident in two samples with a small proportion in a third and associated with underlying UC-PSC (Table 1). The last case of PSC-iCCA was in a 72 F without a history of inflammatory bowel disease (IBD). The diagnosis of PSC was made at the time of liver biopsy for CCA and confirmed in the pathology of normal liver at resection. In this case SBS1 was dominant.
Of the three cases of UC-PSC who were all ≤45 years, each presentation of CCA also led to the diagnosis of PSC either at pathological or radiological review. All three patients had elevation in alpha fetoprotein (AFP) together with carbohydrate antigen 19-9 (CA19-9). Patient 1 was a 40-year-old female with a 15-year history of UC treated with 5-aminosalicyclic acid. Following resection of a T2b tumour she had early recurrence, initially treated with gemcitabine. Thereafter she received cisplatin/gemcitabine for metastatic disease with transient response. The dominant signature in this patient was SBS1 with contributions from SBS8, 9, and 17 (Table 1). Patient 2 was a 26-year-old male presented with jaundice and a locally advanced tumour (Figure 2C). The patient had a prior subtotal colectomy followed by proctectomy and had been on long-standing antibiotics for chronic pouchitis. The final patient also presented with jaundice and a locally advanced tumour at the age of 45 years. This patient had >10-year history of UC and also a concurrent diagnosis of hypereosinophilic syndrome and had been receiving imatinib. The latter two patients progressed rapidly through first line cisplatin/gemcitabine given to downstage their disease and died within 3 months of diagnosis. The dominant signatures in patient 2 were SBS28 and SBS17. This patient also had an elevated TMB at 12 Mut/Mb. The third patient’s tumour expressed a mix of mutational signatures, predominantly SBS9, SBS8, SBS2, SBS13, and SBS1 with a small contribution from SBS17.
Discussion
Patients with advanced cancer with cirrhosis and /or autoimmune disorders are under-represented in clinical trials (28). Here, we demonstrate that in such patients, who often have shorter survival, TP53 mutations underscore this aggressive cohort. In addition, we observed a significant enrichment of SBS17 in those with underlying UC-PSC, although our numbers are small and this can only be considered hypothesis generating. While a recent large study exploring a 42 gene panel in PSC-CCA documents a predominance of TP53, SMAD4, KRAS, and CDKN2A alterations (18), few studies have performed WGS in CCA patients (15). We confirm previous findings showing that PSC-CCA contain molecular characteristic more in keeping with extrahepatic cholangiocarcinoma (eCCA) (18). Our study suggests that KRAS, TP53, CDKN2A, and SMAD4 mutations are enriched in aggressive CCA. Additionally, we observed multiple mutational signatures across CCA samples. Similar to reports by the International Cancer Genome Consortium, SBS1 and SBS5 dominate (15). We note age related SBS5 to be more prevalent in LTS whereas STS harbour a greater number of mutations associated with SBS1, the clock-like mutational signature (29). Our limited dataset detected potential FGFR2 fusions, a finding characteristic of SD iCCA, exclusively in long term survivors while STSs possessed genetic alterations characteristic of LD iCCA, pCCA, and eCCA.
Prior genetic analysis identified unique epigenetic changes in 2 molecular clusters of CCA (15). Interestingly, Clusters 1 and 4 were identified as two unique hypermethylated groups. On the contrary, Clusters 2 and 3 displayed low levels of methylation. Though the patterns of hypermethylation were different between Cluster 1 and 4 and different gene promoters were targeted, Gene-Set Enrichment Analysis revealed the alteration of similar downstream pathways. Different mechanisms of hypermethylation were identified between Clusters 1 and 4. Cluster 1 had diminished expression of the demethylation enzyme TET1 and increased expression of the histone methyltransferase EZH2. Cluster 4 was enriched in IDH1/2 and BAP1 mutations. Both BAP1 and IDH1/2 mutations have been associated with hypermethylation in CCA. Of note, frequent mutations in the chromatin remodelling genes BAP1, ARID1A, and PBRM1 have previously been described in iCCA and these genes were frequently mutated in our dataset as well. BAP1 mutations were found in 5/20 samples and were equally distributed between iCCA and pCCA. ARID1A mutations were identified in 2/20 samples, both were iCCA characterized by STS. PBRM1 mutations were identified in 2/20 samples. Though the prognostic role of these chromatin remodelling genes is uncertain, they are likely to have an important role in CCA development and given the extensive epigenetic changes seen in molecular subgroups of CCA, epigenetic targeted therapy may represent a therapeutic intervention in specific subgroups.
SBS17 was previously reported across clusters 1–3 (Fluke associated, TP53 enriched, and immunogenic) in small proportions in the International Cancer Genomics Consortium (ICGC) dataset where only 1% of PSC-CCA were included (15). It has been shown to associate with prior 5-fluorouracil (5-FU) treatment, esophageal adenocarcinoma, and possibly induced by acid reflux and reactive oxidative species (30-32). The identification of SBS17 in PSC patients may hold important clinical implications. CCA is an extraordinarily lethal disease when occurring in patients with PSC. Notably, CCA is responsible for one third of the all-cause mortality in PSC patients and 72% of PSC patients who develop CCA will die within one year (33). Patients with PSC have a 20% lifetime incidence of developing CCA; a 400- to 500-fold higher risk when compared to the general population. Despite this, the role of surveillance remains controversial. There is little evidence to support an effective screening strategy and the detection of CCA in PSC remains difficult due to significant abnormalities already present throughout the biliary tree (33). Despite this, the American Gastroenterological Association recommends surveillance for all PSC patients every 6 to 12 months involving computed tomography (CT), magnetic resonance imaging (MRI), or ultrasound (US) with or without CA19-9 (34). Other organization such as the European Association for Study of Liver Disease recommend against any routine testing for the early detection of CCA. Notably, the majority of PSC patients will not develop CCA and a better method is required to identify those PSC patients who are at higher risk of CCA development, in whom, an intense screening program may be beneficial. The detection of SBS17 in PSC biopsies may identify those patients at high risk of malignant transformation. Though the specific aetiology of SBS17 remains unknown, it is thought to be related to oxidative DNA damage (31). SBS17-associated mutations have been identified in Barrett’s Oesophagus, the pre-malignant condition of oesophageal adenocarcinoma (35). It is absent from normal oesophageal samples. Interestingly, single cell DNA Sequencing showed a unique association with SBS17-mutations and chromosomal instability (CIN) in Barret’s Oesophagus. SBS17 was only identified in those cells that possessed CIN and absent from chromosomally stable cells (35). It is plausible that SBS17-associated mutations may be early oncological events in patients with PSC and these mutations may provide a causal link between PSC and the development of CCA. Certainly, additional studies are warranted to identify the significance of SBS17 in the development of CCA in patients with PSC. The detection of SBS17 could identify those high-risk patients where intense screening is warranted. Additionally, mutational signature could be obtained from whole exome sequencing of brush cytology samples and the identification of SBS17 in PSC patients with equivocal brush cytology could improve the specificity and sensitivity of diagnosis. Notably, studies have sought to explore KRAS mutation detection in the bile of patients with PSC as a screening mechanism for CCA but have failed to show benefit (36). In addition to the detection of high-risk genetic signatures, our study would suggest that TP53 inactivation could be an additional screening marker in this patient cohort.
Our study identified 3 patients with significantly elevated TMB, all within the STS group. Notably, the Food and Drug Administration (FDA) has approved pembrolizumab for treatment of adult patients with metastatic solid tumours with TMB >10 mut/mb (37). The recent TOPAZ-1 clinical trial, evaluating Durvalumab in patients with advanced biliary tract cancer displayed a significant improvement in OS, progression-free survival (PFS), and objective response rate (ORR) in patients treated with durvalumab (38). However, programmed death ligand 1 (PDL1) was not predictive of response to immunotherapy and though a percentage of patients clearly benefited from immunotherapy-based treatment, biomarkers are required. Other studies investigating pembrolizumab in patients with advanced CCA have revealed durable responses in subsets of patients which are again, independent of PDL1 status (39). Our work did not interrogate the immunophenotypes of CCA, however there is an urgent need to identify predictive biomarkers for immunotherapy. Previous molecular analyses highlight an important immunoregulatory role in subsets of CCA (15,40).
Compared to traditional methods, WGS provides a more extensive analysis of the comprehensive mutational landscape occurring in tumour samples. As opposed to whole-exome sequencing (WES) or targeted sequencing, WGS allows the analysis of all portions of the DNA including coding, non-coding, and mitochondrial DNA. It also allows for the identification of a greater breath of variants including SNVs, indels, structural variants, and copy-number variants. Notably, adequate tissue sampling remains a challenge for WGS and limits its applications to samples with low cellularity such as brush cytology. To ensure the accuracy and reliability of WGS data, all the sequencing performed at our institution undergoes LCM to ensure sufficient cellularity and rigorous quality control (QC) metrics.
This study has several limitations. The study is retrospective and includes a small number of patients. The cases were selected based on availability of tissue and survival but without matched analyses. We however provide additional WGS mutational data which are limited in the field. We did not have RNA sequencing data to validate putative fusions or provide subtyping information and methylation data was not available.
Conclusions
In conclusion, WGS provides additional biological information regarding the heterogenous nature of CCA particularly in patients with underlying inflammatory disorders, where TP53 mutations are prevalent and mutational signatures may be unique. Ongoing prospective observational studies at our institution will seek to validate these findings.
Acknowledgments
Funding: This study was generously funded by the Marie Thompson Fund with additional support from the Legresley Biliary Fund, through the Princess Margaret Cancer Foundation.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-676/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-676/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-676/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-676/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kirstein MM, Vogel A. Epidemiology and Risk Factors of Cholangiocarcinoma. Visc Med 2016;32:395-400. [Crossref] [PubMed]
- Banales JM, Marin JJG, Lamarca A, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol 2020;17:557-88. [Crossref] [PubMed]
- Chung T, Park YN. Up-to-Date Pathologic Classification and Molecular Characteristics of Intrahepatic Cholangiocarcinoma. Front Med (Lausanne) 2022;9:857140. [Crossref] [PubMed]
- Ma B, Meng H, Tian Y, et al. Distinct clinical and prognostic implication of IDH1/2 mutation and other most frequent mutations in large duct and small duct subtypes of intrahepatic cholangiocarcinoma. BMC Cancer 2020;20:318. [Crossref] [PubMed]
- Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003-10. [Crossref] [PubMed]
- Wang XY, Zhu WW, Wang Z, et al. Driver mutations of intrahepatic cholangiocarcinoma shape clinically relevant genomic clusters with distinct molecular features and therapeutic vulnerabilities. Theranostics 2022;12:260-76. [Crossref] [PubMed]
- Primrose JN, Fox RP, Palmer DH, et al. Capecitabine compared with observation in resected biliary tract cancer (BILCAP): a randomised, controlled, multicentre, phase 3 study. Lancet Oncol 2019;20:663-73. [Crossref] [PubMed]
- Gringeri E, Gambato M, Sapisochin G, et al. Cholangiocarcinoma as an Indication for Liver Transplantation in the Era of Transplant Oncology. J Clin Med 2020;9:1353. [Crossref] [PubMed]
- Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273-81. [Crossref] [PubMed]
- Shroff RT, Javle MM, Xiao L, et al. Gemcitabine, Cisplatin, and nab-Paclitaxel for the Treatment of Advanced Biliary Tract Cancers: A Phase 2 Clinical Trial. JAMA Oncol 2019;5:824-30. [Crossref] [PubMed]
- Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin Cancer Res 2018;24:4154-61. [Crossref] [PubMed]
- Lee DH, Lee JM. Primary malignant tumours in the non-cirrhotic liver. Eur J Radiol 2017;95:349-61. [Crossref] [PubMed]
- Khan SA, Tavolari S, Brandi G. Cholangiocarcinoma: Epidemiology and risk factors. Liver Int 2019;39:19-31. [Crossref] [PubMed]
- Clements O, Eliahoo J, Kim JU, et al. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: A systematic review and meta-analysis. J Hepatol 2020;72:95-103. [Crossref] [PubMed]
- Jusakul A, Cutcutache I, Yong CH, et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov 2017;7:1116-35. [Crossref] [PubMed]
- Ahrendt SA, Pitt HA, Nakeeb A, et al. Diagnosis and management of cholangiocarcinoma in primary sclerosing cholangitis. J Gastrointest Surg 1999;3:357-67; discussion 367-8. [Crossref] [PubMed]
- Churi CR, Shroff R, Wang Y, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 2014;9:e115383. [Crossref] [PubMed]
- Goeppert B, Folseraas T, Roessler S, et al. Genomic Characterization of Cholangiocarcinoma in Primary Sclerosing Cholangitis Reveals Therapeutic Opportunities. Hepatology 2020;72:1253-66. [Crossref] [PubMed]
- McNamara MG, Walter T, Horgan AM, et al. Outcome of adjuvant therapy in biliary tract cancers. Am J Clin Oncol 2015;38:382-7. [Crossref] [PubMed]
- Notta F, Chan-Seng-Yue M, Lemire M, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 2016;538:378-82. [Crossref] [PubMed]
- Connor AA, Denroche RE, Jang GH, et al. Association of Distinct Mutational Signatures With Correlates of Increased Immune Activity in Pancreatic Ductal Adenocarcinoma. JAMA Oncol 2017;3:774-83. [Crossref] [PubMed]
- Connor AA, Denroche RE, Jang GH, et al. Integration of Genomic and Transcriptional Features in Pancreatic Cancer Reveals Increased Cell Cycle Progression in Metastases. Cancer Cell 2019;35:267-282.e7. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28:1811-7. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017;23:517-25. [Crossref] [PubMed]
- Piñero F, da Fonseca LG. Trial eligibility in advanced hepatocellular carcinoma: Does it support clinical practice in underrepresented subgroups? World J Gastroenterol 2021;27:3429-39. [Crossref] [PubMed]
- Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012;149:979-93. [Crossref] [PubMed]
- Pich O, Muiños F, Lolkema MP, et al. The mutational footprints of cancer therapies. Nat Genet 2019;51:1732-40. [Crossref] [PubMed]
- Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94-101. [Crossref] [PubMed]
- Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 2013;45:478-86. [Crossref] [PubMed]
- Fung BM, Tabibian JH. Cholangiocarcinoma in patients with primary sclerosing cholangitis. Curr Opin Gastroenterol 2020;36:77-84. [Crossref] [PubMed]
- Bowlus CL, Lim JK, Lindor KD. AGA Clinical Practice Update on Surveillance for Hepatobiliary Cancers in Patients With Primary Sclerosing Cholangitis: Expert Review. Clin Gastroenterol Hepatol 2019;17:2416-22. [Crossref] [PubMed]
- Busslinger GA, de Barbanson B, Oka R, et al. Molecular characterization of Barrett's esophagus at single-cell resolution. Proc Natl Acad Sci U S A 2021;118:e2113061118. [Crossref] [PubMed]
- Kubicka S, Kühnel F, Flemming P, et al. K-ras mutations in the bile of patients with primary sclerosing cholangitis. Gut 2001;48:403-8. [Crossref] [PubMed]
- Marcus L, Fashoyin-Aje LA, Donoghue M, et al. FDA Approval Summary: Pembrolizumab for the Treatment of Tumor Mutational Burden-High Solid Tumors. Clin Cancer Res 2021;27:4685-9. [Crossref] [PubMed]
- Oh DY, Lee KH, Lee DW, et al. Gemcitabine and cisplatin plus durvalumab with or without tremelimumab in chemotherapy-naive patients with advanced biliary tract cancer: an open-label, single-centre, phase 2 study. Lancet Gastroenterol Hepatol 2022;7:522-32. [Crossref] [PubMed]
- Piha-Paul SA, Oh DY, Ueno M, et al. Efficacy and safety of pembrolizumab for the treatment of advanced biliary cancer: Results from the KEYNOTE-158 and KEYNOTE-028 studies. Int J Cancer 2020;147:2190-8. [Crossref] [PubMed]
- Lin J, Dai Y, Sang C, et al. Multimodule characterization of immune subgroups in intrahepatic cholangiocarcinoma reveals distinct therapeutic vulnerabilities. J Immunother Cancer 2022;10:e004892. [Crossref] [PubMed]


