
This article has an erratum available at: http://dx.doi.org/10.21037/jgo-23-392 the article has been update on 2023-05-05 at here.
MicroRNA-543 controls pancreatic cancer development by LINC00847-microRNA-543-STK31 axis
Highlight box
Key findings
• microRNA-543, through the LINC00847/microRNA-543/STK31 axis, plays a role in the development of PC as a tumor suppressor.
What is known and what is new?
• MicroRNA-543 has been reported to control the occurrence and development of various cancers, such as cervical cancer, ovarian cancer, colorectal cancer, and non-small cell lung cancer, among others
• The role of miR-543 in pancreatic cancer is less known. Then we research the functions of miR-543 by ceRNA network in Pancreatic cancer cells and tissues.
What is the implication, and what should change now?
• How miR-543 plays a role in pancreatic cancer deserves our study, and it may become a target for diagnosis and prognosis of pancreatic cancer.
Introduction
Pancreatic cancer (PC) is one of the most deadly types of gastrointestinal tract cancer. It is usually recognized at an advanced stage and has a poor prognosis. The 5-year survival rate is less than 10% (1,2). PC is still treated with traditional methods, and only about 20% of patients have an opportunity to undergo surgery, leaving the rest with conservative treatment (3). Recently, targeted therapy and immunotherapy have gradually become the focus of research (4). However, the study of targeted immunity in PC is not very thorough. Therefore, targeted molecular markers are needed for the diagnosis and treatment of PC.
MicroRNA-543 has been reported to control the occurrence and development of various cancers, such as cervical cancer, ovarian cancer, colorectal cancer, and non-small cell lung cancer, among others (5-8). However, less is known about its role in PC. Serine/threonine kinase 31 (STK31) controls tumorigenicity via control of differentiation states, suggesting that STK31 may be controlled by an epigenetic mechanism (9). Recent research reported that STK31 controlled the cell growth and cell cycle of lung cancer cells via the Wnt/beta-catenin pathway (10), maintained the undifferentiated state of colon cancer cells (9), and was a novel biomarker of colorectal cancer (11). Interestingly, there are few studies on the mechanism of STK31 in PC.
Long noncoding RNAs (lncRNAs) are defined as functional gene transcripts that are longer than 200 nucleotides and lack protein-coding capacity (12). A large body of research has found that lncRNAs are engaged in a variety of physiological events, such as the immune system response, inflammatory processes, and the advancement of cancer (13-15). There is increasing evidence that abnormal control of lncRNAs is closely related to the development of cancer, which can be linked to their carcinogenic or anticancer properties (16). LINC00847 has been reported to be involved in apoptotic pathways in kidney cancer (17), contributes to hepatocellular carcinoma development (18), and was shown to be induced by E2F1 to accelerate non-small cell lung cancer development by targeting the miR-147a/IFITm1 axis (19). Despite this, very little is known about its function in PC and the underlying mechanisms that cause the disease.
In this study, bioinformatics was utilized with experimental testing, which found that the abnormal transcription of microRNA-543 may affect the development of PC. As a result of our research, we observed that the tumor promoter LINC00847 has therapeutic significance since it sponges microRNA-543 to target the STK31 gene. Therefore, the LINC00847/microRNA-543/STK31 signaling axis may be a promising therapeutic target for PC treatment and a novel marker for the diagnosis of PC. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1017/rc).
Methods
Patient tissue samples and ethics statement
Human PC tissues and adjacent tissues were obtained from patients admitted to China-Japan Union Hospital of Jilin University from March 2020 to January 2022. All tissue specimens were stored at −80 ℃ until use. The study was conducted according to the Helsinki Declaration (as revised in 2013) and approval was obtained from the Ethics Committee of China-Japan Union Hospital of Jilin University (No. 20221014001). Before the beginning of the study, each patient signed written informed consent.
Transfection and cell culture
The human PC cell lines BxPC-3, SW1990, and PANC-1 were acquired from Cell Resource Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China). The human PC cell line CFPAC-1 was purchased from Kunming Cell Bank, Chinese Academy of Sciences (Kunming, China). The human normal pancreatic cell line H6C7 was purchased from Shanghai Yaji Biological. BxPC-3 cells were cultured in RPMI 1640 medium (Gibco, USA). H6C7, CFPAC-1, and PANC-1 cells were cultured in DMEM medium (Gibco, USA). SW1990 cells were cultured in L-15 Leibovitz Medium. All cell mediums were supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) at 37 ℃ in a humidified atmosphere containing 5% CO2.
Vectors encoding STK31 (oe-STK31), miRNA inhibitor (microRNA-543 inhibitor), negative control (NC vector), control mimic, miRNA mimic (microRNA-543 mimic), control inhibitor, LINC00847 overexpression (oe-LINC00847), and the overexpression control (vector-NC) lentiviruses for LINC00847 silencing (si-LINC00847) and the silencing control (si-NC) were purchased from Ribobio (Guangzhou, China). The manufacturer’s procedure was followed for transfection and infection of cells.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells using TRIzol (10296010, Invitrogen, USA) reagent. Reverse transcriptions were performed using the PrimeScript RT Master Mix (code No. RR036A, Takara, China) with random primers for the mRNAs and lncRNAs. Reverse transcriptions were performed using the Mir-X miRNA First-Strand Synthesis Kit (code No. 638315, Takara, China) with specific stem-loop primers for microRNA-543. Subsequently, qRT-PCR reaction was performed using TB Green Premix Ex Taq II (code No. RR820Q/A/B, Takara, China) and Mir-X miRNA qRT-PCR TB Green® Kit (code No. 638314, Takara, China) with an ABI Prism 7500 sequence detection system (Prism® 7500, Applied Biosystems, USA). β-actin and U6 were used as internal controls. The transcription of microRNA-543, STK31, and LINC00847 was quantified by the 2-ΔΔCt method. The primers used in the study are shown in Table 1.
Table 1
| Target gene | Forward/reverse | Primer sequences |
|---|---|---|
| β-Actin | Forward | 5'-CATGTACGTTGCTATCCAGGC-3' |
| Reverse | 5'-CTCCTTAATGTCACGCACGAT-3' | |
| U6 | Forward | 5'-CTCGCTTCGGCAGCACA-3' |
| Reverse | 5'-AACGCTTCACGAATTTGCGT-3' | |
| microRNA-543 | Forward | 5'-AAACATTCGCGGTGCGCGTAC-3' |
| Reverse | 5'-GCAGGGTCCGAGGTATTC-3' | |
| STK31 | Forward | 5'-GTCAAGTGTCTGCAAAGAGCTGG-3' |
| Reverse | 5'-CTGTGGCAATGAGCCTTTCCTC-3' | |
| LINC00847 | Forward | 5'-CGGCTGGAGCGAAGAGT-3' |
| Reverse | 5'-GAGAACCGCAGAAGAACG-3' '' |
Western blotting (WB)
The cell samples were lysed using RIPA buffer (purchased from Beyotime, P0013B, China), which was mixed with a phosphatase inhibitor cocktail. Sodium Dodecyl Sulfate PolyAcrylamide Gel Electrophoresis (SDS-PAGE) was utilized to isolate the proteins, which were then deposited to polyvinylidene fluoride (PVDF) membranes for further examination and investigation. After blocking with skimmed milk (5%), primary antibodies directed against specific proteins of interest were incubated over PVDF membranes for a short period of time before being utilized. Antibodies were acquired from Abcam in Cambridge, MA (USA) and used at a 1:1,000 dilution for STK31 (ab155172), Slug (ab27568), Snail (ab216347), β-actin (ab8226), and TWIST (ab50887, 0.2–2 µg/mL).
N-cadherin (13116), E-cadherin (14472), and Vimentin (46173) antibodies were bought from Cell Signaling Technology (CST, Danvers, MA, USA) and used at a 1:1,000 dilution. A secondary antibody comprising horseradish peroxidase (HRP, ab205719) was then administered after the sample was washed several times. Lastly, the protein bands were observed using the EasyBlot ECL kit (Sangon, China).
Cell growth assay
Stably transfected PC cells were cultured in 96-well plates (1×104 cells per well). Cell viability was assessed using the CCK-8 (C0037, Beyotime, China) assay. The absorbance at 450 nm was then measured with a microplate reader. Growth in the number of novel PC cells was also determined by performing a colony formation experiment (1×104 cells per well of 6-well plates).
Transwell assay
For the purpose of monitoring invasion and metastasis, Transwell migration chambers were utilized (24-well chambers, 354572 Corning, USA). Matrigel (356234 BD, Solarbio, China) was applied to the container inserts prior to cell invasion trials. In both tests, the upper chamber of each well was seeded with 2×105 cells in 300 µL of serum-free media. A mixture of 600–800 µL DMEM (E600003, Sangon Biotech) supplemented with 10% FBS (E510008, Sangon Biotech) was used to fill the lower chambers of the Transwell. Subsequently, the cells were incubated for 24 hours. The top chambers were then rinsed with phosphate-buffered saline (PBS, E607008, Sangon Biotech), fixed with 4% paraformaldehyde (PFA, E672002, Sangon Biotech) for 30–60 minutes, and dyed with 1% crystal violet (A600331, Sangon Biotech) to obtain the desired appearance. Photomicrographs of the cell layers were taken with an Olympus BX51 microscope.
Flow cytometry (FCM) analysis
PC cells were digested with trypsin (A003702, Sangon Biotech) without EDTA and washed twice with cold PBS and collected by centrifugation. Then, 5 µL Annexin V-FITC (E606336, Sangon Biotech) and propidium iodide (E606336, Sangon Biotech) were added to the cells (1×106 cells/mL) and mixed well, then placed at room temperature without light for 15 minutes. After incubation, 200 µL of 1× binding buffer (E606336, Sangon Biotech) was added, then FCM was utilized to detect apoptosis within 1 hour.
Dual-luciferase reporter gene assay
We generated and cloned a sequence including the putative binding site of LINC00847 and the 3′UTR of STK31 as well as the corresponding mutant sequence (GenePharma, Shanghai, China) into the pGL3 luciferase reporter vector. Lipofectamine 2000 (11668019, Invitrogen, USA) was utilized for all co-transfection assays. The co-transfection of BxPC-3 and PANC-1 cells with transcription plasmids and either a microRNA-543 mimic or a control mimic was carried out. At 24 hours after transfection, evaluation of relative luciferase activity was performed using the Nano-Glo® Dual-Luciferase Reporter kit (E1910, Promega, USA) in accordance with the manufacturer’s instructions.
RNA immunoprecipitation (RIP) assay
The RIP assay was carried out in accordance with the manufacturer’s instructions using the Magna RIP assay kit, which was purchased from Applied Biosystems (17-704, Sigma-Aldrich). RIP lysis buffer was applied to BxPC-3 and PANC-1 cells prior to the addition of magnetic beads containing control IgG antibodies (17-704, Sigma-Aldrich). Next, RT-qPCR was performed to identify the most abundant RNA fragments.
RNA pull-down assay
GenePharma (Shanghai, China) provided the biotinylated microRNA-543, and mutant and NC were utilized as controls. After the oligonucleotides were synthesized, the lysates from BxPC-3 and PANC-1 cells were treated with M-280 streptavidin magnetic beads (11206D, Invitrogen, USA) for 30 minutes. Subsequently, we extracted the RNA bound to STK31/LINC00847 and performed quantitative real-time PCR to determine its concentration.
RNA fluorescence in-situ hybridization (FISH)
FISH was carried out using the RiboTM FISH kit (Ribobio, China) in reference to previous research steps (20). The probes of LINC00847 were synthesized and modified by RiboBio (Guangzhou, China), and cellular DNA was stained with 4',6-diamidino-2-phenylindole (DAPI) and photographed by confocal microscopy.
Bioinformatics analysis
MicroRNA-543 was found to have abnormal transcription and was associated with poor prognosis in 160 PC patients from the The Cancer Genome Atlas (TCGA) database (21). The StarBase 3.0 (http://starbase.sysu.edu.cn) database and Kaplan-Meier plotter (http://kmplot.com/analysis/index.php?p=service) also showed the same results. Furthermore, RNA22 (https://cm.jefferson.edu/rna22/) and microRNA.org (http://www.microrna.org/) predicted that microRNA-543 interacted with STK31, and previous research also reported that microRNA-543 plays a role in PC by targeting STK31 (22). Then, we selected STK31 as a target gene of microRNA-543 for further research. The probable target lncRNAs of microRNA-543 were identified using Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/) and the Starbase3.0 and LncBase (http://carolina.imis.athena-innovation.gr) databases. The transcription of lncRNAs was evaluated by GEPIA2 (http://gepia2.cancer-pku.cn/#analysis).
Statistical analysis
The data (mean ± SD) were evaluated by SPSS 22.0 and GraphPad Prism 9. Student’s t-tests were employed to establish whether or not the data was statistically significant. Kaplan-Meier survival curves were utilized to examine the link between microRNA-543 level and PC patient survival. The Kaplan-Meier survival curves were used to analyze the survival distributions, and the significance of the differences between 2 groups was determined using the log-rank test for the microRNA-543 levels in PC tissue samples. The corresponding graphs were drawn with GraphPad Prism 9, and *&#, P<0.05, **&##, P<0.01, or ***&###, P<0.001 were considered statistically significant.
Results
MicroRNA-543 is downregulated in PC tissues and cells and is associated with poor prognosis
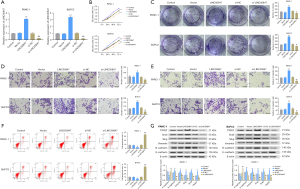
MicroRNA-543 was found to have low expression in 160 PC patients using TCGA database (21). Using the StarBase and Kaplan-Meier plotter databases, we also found that microRNA-543 was downregulated in PC tissues and was associated with poor survival (Figure 1A,1B). Furthermore, microRNA-543 was obviously downregulated in PC tissues and cells compared with normal tissues and H6C7 cells (Figure 1C,1D, P<0.001). We also collected the clinical data of PC patients and it was found that the expression level of microRNA-543 was significantly correlated with T stage (P<0.05, Table 2).
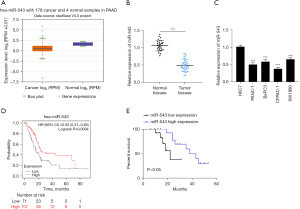
Table 2
| Characteristics | Case | microRNA-543 expression | P value | |
|---|---|---|---|---|
| Low (n=20) | High (n=10) | |||
| Age at surgery (years) | 0.7888 | |||
| <60 | 11 | 7 | 4 | |
| ≥60 | 19 | 13 | 6 | |
| Gender | 0.1927 | |||
| Male | 17 | 13 | 4 | |
| Female | 13 | 7 | 6 | |
| Tumor size (cm) | 0.4386 | |||
| ≥3 | 15 | 11 | 4 | |
| <3 | 15 | 9 | 6 | |
| T grade | 0.0371* | |||
| T1+T2 | 13 | 6 | 7 | |
| T3+T4 | 17 | 14 | 3 | |
| Lymph node invasion | 0.7842 | |||
| Negative (N0) | 20 | 13 | 7 | |
| Positive (N1-N3) | 10 | 7 | 3 | |
| TNM stage | 0.5921 | |||
| I-II | 19 | 12 | 7 | |
| III-IV | 11 | 8 | 3 | |
| Histological grade | 0.0701 | |||
| Low | 16 | 13 | 3 | |
| Middle-high | 14 | 7 | 7 | |
*, P<0.05. TNM, tumor node metastasis.
Additionally, the Kaplan-Meier method was used to generate overall survival (OS) curves, and survival information of previously registered patients was gathered. Patients who had low levels of microRNA-543 expression in PC tissues had a significantly shorter OS rate (P<0.05, Figure 1E). These data indicated that microRNA-543 may be a novel marker in the development of PC.
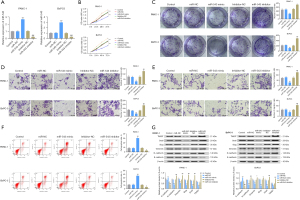
Knockdown of microRNA-543 promotes the development and epithelial-mesenchymal transition of PC in vitro
In order to study the molecular function of microRNA-543 in PC cells, BxPC-3 and PANC-1 cells were selected to construct stable cell lines to evaluate how microRNA-543 affects the development of PC. As shown in Figure 2A, the stable transfection of cell lines was confirmed compared with the control group. CCK-8 and colony formation assays were performed to investigate the growth of PC cells. The results revealed that microRNA-543 silencing promoted the growth of PC cells, while microRNA-543 overexpression suppressed the growth of PC cells compared with controls (Figure 2B,2C). In addition, the Transwell assay was performed to evaluate the impact of microRNA-543 expression on the invasion and metastasis of PC cells. From the results, we found that microRNA-543 knockdown substantially enhanced the invasion and migration of PC cells compared with the controls. On the other hand, overexpression of microRNA-543 slowed cell invasion and migration dramatically (Figure 2D,2E). Furthermore, the FCM assay was performed to analyze the role of microRNA-543 in the apoptosis PC cells. We found that microRNA-543 silencing substantially inhibited cell apoptosis compared with controls, whereas overexpression of microRNA-543 substantially enhanced cell apoptosis (Figure 2F). Finally, EMT marker expression, a key biological process of tumor cell migration and invasion (23), was detected by WB. Interestingly, the results showed that knockdown of microRNA-543 enhanced N-cadherin, Vimentin, Slug, Snail1, and TWIST protein expression and inhibited E-cadherin protein expression in BxPC-3 and PANC-1 cells, indicating a shift from an epithelial to mesenchymal phenotype. When microRNA-543 was increased, PC cells were unable to acquire the EMT phenotype (Figure 2G).
MicroRNA-543 controls PC development by targeting STK31
To further explore the function of microRNA-543, we used published bioinformatics articles and online software prediction and found that microRNA-543 acts by targeting STK31 (22). In our study, we further verified the relationship between microRNA-543 and STK31. As shown in Figure 3A, we found that STK31 is highly expressed in PC tissues compared with normal tissues. Using qRT-PCR and WB assays, we also found that STK31 expression was significantly higher in BxPC-3 and PANC-1 cells than in H6C7 cells (Figure 3B,3C). Then, we transfected microRNA-543 inhibitor and microRNA-543 mimics into BxPC-3 and PANC-1 cell lines and found that the transcription of STK31 was decreased when microRNA-543 was overexpressed, while the transcription of STK31 was increased when microRNA-543 was knocked down (Figure 3D). The predicted binding site for microRNA-543 and STK31 is shown in Figure 3E. The interaction was verified by a luciferase reporter assay. A decrease in luciferase activity was seen in BxPC-3 and PANC-1 cells expressing the wild-type (WT) STK31 reporter, whereas the luciferase activity in cells expressing the mutant (MUT) STK31 reporter was unaffected (Figure 3E). The direct interaction between STK31 and microRNA-543 was further confirmed by RIP and RNA-pull down assays, which demonstrated that STK31 directly binds to microRNA-543 (Figure 3F,3G). Thus, we selected STK31 for further study. Then, the relationship between STK31 expression and the clinical characteristics of PC patients were listed in Table 3. The expression of STK31 was significantly correlated with histological grade (P=0.0173).
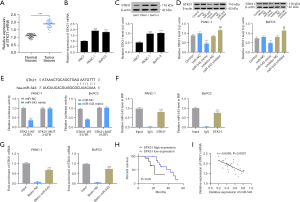
Table 3
| Characteristics | Case | STK31 expression | P value | |
|---|---|---|---|---|
| Low (n=11) | High (n=19) | |||
| Age at surgery (years) | 0.4473 | |||
| <60 | 11 | 5 | 6 | |
| ≥60 | 19 | 6 | 13 | |
| Gender | 0.0877 | |||
| Male | 17 | 4 | 13 | |
| Female | 13 | 7 | 6 | |
| Tumor size (cm) | 0.7048 | |||
| ≥3 | 15 | 6 | 9 | |
| <3 | 15 | 5 | 10 | |
| T grade | 0.0877 | |||
| T1+T2 | 13 | 7 | 6 | |
| T3+T4 | 17 | 4 | 13 | |
| Lymph node invasion | 0.1804 | |||
| Negative (N0) | 20 | 9 | 11 | |
| Positive (N1-N3) | 10 | 2 | 8 | |
| TNM stage | 0.9791 | |||
| I-II | 19 | 7 | 12 | |
| III-IV | 11 | 4 | 7 | |
| Histological grade | 0.0173* | |||
| Low | 16 | 9 | 7 | |
| Middle-high | 14 | 2 | 12 | |
*, P<0.05. TNM, tumor node metastasis.
The Kaplan-Meier survival curve also showed an inverse correlation between the expression level of STK31 and the prognosis of PC patients. Specifically, PC patients with high STK31 expression had a shorter OS (Figure 3H). Furthermore, results of qRT-PCR from PC tissues showed a highly negative correlation between microRNA-543 and STK31 expression (Figure 3I). Therefore, the results suggest that STK31 may be the target gene of microRNA-543 and a prognostic marker of PC.
LINC00847 regulates the tumor development of PC and EMT in vitro by absorbing microRNA-543
To further investigate the novel function of microRNA-543, LncBase and starBase3.0 were utilized as bioinformatics tools to find possible lncRNA targets of microRNA-543. As shown in Figure 4A, 11 lncRNAs were obtained. Then, through comprehensive analysis and literature research, LINC00847 was selected as the target molecule, which was highly expressed in PC (Figure 4B). qRT-PCR analysis revealed that LINC00847 was obviously expressed in PC tissues and cells compared with normal tissues and H6C7 cells (Figure 4C,4D). Furthermore, when we collected the clinical data of the PC patients, it was found that the expression level of LINC00847 significantly correlated with the TNM stage of PC tissues (P=0.0344, Table 4).
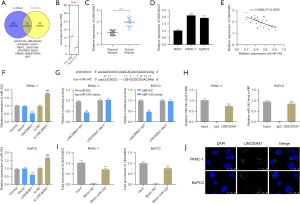
Table 4
| Characteristics | Case | LINC00847 expression | P value | |
|---|---|---|---|---|
| Low (n=13) | High (n=17) | |||
| Age at surgery (years) | 0.5578 | |||
| <60 | 11 | 4 | 7 | |
| ≥60 | 19 | 9 | 10 | |
| Gender | 0.2246 | |||
| Male | 17 | 9 | 8 | |
| Female | 13 | 4 | 9 | |
| Tumor size (cm) | 0.7125 | |||
| ≥3 | 15 | 6 | 9 | |
| <3 | 15 | 7 | 8 | |
| T grade | 0.7851 | |||
| T1+T2 | 13 | 6 | 7 | |
| T3+T4 | 17 | 7 | 10 | |
| Lymph node invasion | 0.1927 | |||
| Negative (N0) | 20 | 7 | 13 | |
| Positive (N1-N3) | 10 | 6 | 4 | |
| TNM stage | 0.0344* | |||
| I-II | 19 | 11 | 8 | |
| III-IV | 11 | 2 | 9 | |
| Histological grade | 0.1269 | |||
| Low | 16 | 9 | 7 | |
| Middle-high | 14 | 4 | 10 | |
*, P<0.05. TNM, tumor node metastasis.
Furthermore, results of qRT-PCR in PC tissues showed a highly negative correlation between microRNA-543 and LINC00847 expression (Figure 4E). Then, we transfected si-LINC00847 and oe-LINC00847 in BxPC-3 and PANC-1 cell lines and found that the transcription of microRNA-543 was decreased when LINC00847 was overexpressed, while the transcription of microRNA-543 was increased when LINC00847 was knocked down (Figure 4F). An assay for luciferase reporter activity was performed to confirm this interaction. BxPC-3 and PANC-1 cells expressing the WT LINC00847 reporter showed decreased luciferase activity in the microRNA-543 overexpression group, whereas cells expressing the MUT LINC00847 reporter showed no change in luciferase activity in the microRNA-543 perturbation group (Figure 4G). RIP and RNA-pull down assays were further applied to confirm the direct interaction between LINC00847 and microRNA-543, which demonstrated that LINC00847 directly binds to microRNA-543 (Figure 4H,4I). FISH assays showed that LINC00847 RNA was concentrated in the cytoplasm of PC cells (Figure 4J). Therefore, we can confirm that LINC00847 is the target gene of microRNA-543.
To further evaluate the function of LINC00847 in PC, BxPC-3 and PANC-1 cells were either transfected with si-LINC00847 to inhibit LINC00847 transcription or transfected with overexpression-LINC00847 to promote LINC00847 transcription. Compared with the control group, cell transfection was successful (Figure 5A). Then, CCK-8 and colony formation assays were utilized to measure the impact of LINC00847 overexpression on the growth of PC cells. We found that overexpression of LINC00847 enhanced PC cell growth in terms of cell number, while silencing LINC00847 inhibited PC cell growth (Figure 5B,5C). Moreover, to assess the invasion and migration of PC cells, the Transwell assay was performed. The results revealed that overexpression of LINC00847 substantially promoted PC cell invasion and migration, while knockdown of LINC00847 had opposite results (Figure 5D,5E). In addition, LINC00847 overexpression suppressed cell apoptosis, while LINC00847 knockdown accelerated cell apoptosis (Figure 5F). Finally, WB was applied to determine how LINC00847 affected the EMT process. According to Figure 5G, higher LINC00847 levels boosted N-cadherin, Vimentin, Slug, Snail, and TWIST protein expression in BxPC-3 and PANC-1 cells while inhibiting E-cadherin protein transcription, indicating the change from an epithelial to mesenchymal phenotype. The capacity of PC cells to develop the EMT feature was decreased through LINC00847 knockdown.
MicroRNA-543 controls the development of PC by the LINC00847/microRNA-543/STK31 axis
Furthermore, we explored whether microRNA-543 plays a carcinogenic role in PC through the LINC00847/microRNA-543/STK31 axis. The rescue experiments were constructed using microRNA-543 inhibitors, oe-STK31, or si-LINC00847. The data obtained from the qRT-PCR and WB assays showed that silencing LINC00847 reduced the transcriptional and translational level of STK31 in BxPC-3 and PANC-1 cells, while the effects of silencing LINC00847 were reversed by microRNA-543 inhibitor or oe-STK31 (Figure 6A,6B).
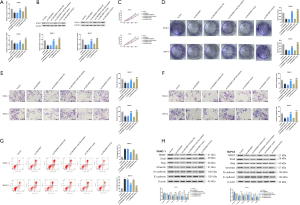
Subsequently, we investigated whether microRNA-543 inhibitor or oe-STK31 might reverse the role of LINC00847 in PC cells. Inhibition of LINC00847 transcription resulted in the decreased cell number, invasion, and migration of PC cells as well as an increase in apoptosis, as determined by the CCK-8, colony formation, Transwell, and FCM assays. The inhibitor of microRNA-543 or oe-STK31 reversed the results (Figure 6C-6G).
WB analysis revealed that the transcription of mesenchymal markers N-cadherin, Snail, and TWIST were all reduced when LINC00847 was silenced; however, when co-transfection with microRNA-543 inhibitor or oe-STK31 was performed, the transcription of these markers was restored. In contrast to the transcription patterns of N-cadherin, Vimentin, Slug, Snail, and TWIST, the transcription pattern of E-cadherin was the inverse of that of the other proteins (Figure 6H). In this study, we hypothesized that microRNA-543 controlled the growth and development of PC cells through the LINC00847/microRNA-543/STK31 axis.
Discussion
PC is dubbed the “king of cancer” due to its aggressive biological nature and a dearth of viable treatments (24,25). As a result, it is critical to identify sensitive therapeutic targets or prognostic indicators to ensure that PC patients are receiving the most effective therapies possible.
We found that microRNA-543 was substantially downregulated and related to survival in PC by TCGA database, which suggested that microRNA-543 might be involved in the development of PC. Liu et al. reported that microRNA-543 inhibited cervical cancer growth and metastasis by targeting TRPM7 (5). Other studies also found that microRNA-543 can control the disease development of cervical cancer, colorectal cancer, non-small lung cancer, and prostate cancer (5,7,8,26). Wang et al. showed that LncRNA PVT1 controls TRPS1 transcription in breast cancer by absorbing microRNA-543 (27). Yuan et al. found that microRNA-543 was associated with the occurrence and prognosis of PC by targeting STK31 through a comprehensive bioinformatics analysis (22). However, they did not perform a thorough study, while we performed a deep study of the mechanism of this result. In our study, we confirmed that microRNA-543 promoted PC cell growth, invasion, migration, and EMT processes by targeting STK31.
Then, using a comprehensive bioinformatics study, we determined that LINC00847 may be the microRNA-543 target molecule. Additionally, we discovered that LINC00847 acted as an oncogene, promoting the cell growth, metastasis, invasion, and EMT of PC cells in vitro. Therefore, we built a competitive endogenous RNA (ceRNA) network of LINC00847/microRNA-543/STK31.
For the purpose of determining whether microRNA-543 promotes tumor progression through the LINC00847/microRNA-543/STK31 axis, rescue tests were designed and carried out using either the microRNA-543 inhibitor, si-LINC00847, or oe-STK31. When qRT-PCR and WB were used to analyze the results, it was discovered that decreasing LINC00847 transcription resulted in higher transcriptional and translational levels of STK31 in BxPC-3 and PANC-1 cells. At the same time, microRNA-543 inhibitors or oe-STK31 were found to be effective in reversing the consequences of decreased LINC00847 transcription.
Furthermore, we explored whether microRNA-543 inhibitors or oe-STK31 could reverse the effects of si-LINC00847 in BxPC-3 and PANC-1 cells, which was found to be effective. LINC00847 was found to inhibit the growth of BxPC-3 and PANC-1 cells, as well as their ability to invade and migrate in the Transwell and FCM assays. This was reversed by the microRNA-543 inhibitor or oe-STK31, which was found to inhibit the cell growth, invasion, and migration of BxPC-3 and PANC-1 cells.
Rescue experiments later indicated that LINC00847 had a negative impact on the oncogenic function of STK31. Also, we observed that decreasing LINC00847 transcription in BxPC-3 and PANC-1 cells resulted in a reduction of STK31 transcription in these cells. The function of STK31, on the other hand, was improved when the microRNA-543 inhibitor or oe-STK31 was added. As a result, our findings showed that LINC00847 acts as a ceRNA by harboring microRNA-543 and, as a result, neutralized the carcinogenic impact of microRNA-543 over the target gene STK31 during the growth of PC. Consequently, microRNA-543 has the potential to be a novel prospective biomarker for PC.
Unfortunately, there are some limitations to this research. Firstly, the validation was only performed in vitro, necessitating further in vivo studies with more clinical samples. Secondly, we only tested the mechanisms in cell lines, though they are incapable of reproducing in vitro or in vivo results in a reliable manner. Primary PC cells extracted from patient tissues or patient-derived xenograft models should be employed for additional validation.
Conclusions
In summary, we identified microRNA-543 as a possible treatment biomarker for PC. Our findings also revealed that LINC00847, and microRNA-543, had the ability to act as a ceRNA, mitigating the antitumor impact of microRNA-543 on the target gene STK31 during the course of PC progression. Our findings therefore suggest that microRNA-543 may be a novel biomarker of the development of PC in humans.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1017/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1017/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1017/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted according to the Helsinki Declaration (as revised in 2013) and approval was obtained from the Ethics Committee of China-Japan Union Hospital of Jilin University (No. 20221014001). Before the beginning of the study, each patient signed written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bear AS, Vonderheide RH, O'Hara MH. Challenges and Opportunities for Pancreatic Cancer Immunotherapy. Cancer Cell 2020;38:788-802. [Crossref] [PubMed]
- Jain T, Dudeja V. The war against pancreatic cancer in 2020 - advances on all fronts. Nat Rev Gastroenterol Hepatol 2021;18:99-100. [Crossref] [PubMed]
- Chen H, Zhuo Q, Ye Z, et al. Organoid model: A new hope for pancreatic cancer treatment? Biochim Biophys Acta Rev Cancer 2021;1875:188466. [Crossref] [PubMed]
- Rocha FG. Landmark Series: Immunotherapy and Targeted Therapy for Pancreatic Cancer. Ann Surg Oncol 2021;28:1400-6. [Crossref] [PubMed]
- Liu X, Gan L, Zhang J. miR-543 inhibites cervical cancer growth and metastasis by targeting TRPM7. Chem Biol Interact 2019;302:83-92. [Crossref] [PubMed]
- Yu Q, Zhang Z, He B, et al. MiR-543 functions as tumor suppressor in ovarian cancer by targeting TWIST1. J Biol Regul Homeost Agents 2020;34:101-10. [PubMed]
- Su DW, Li X, Chen J, et al. MiR-543 inhibits proliferation and metastasis of human colorectal cancer cells by targeting PLAS3. Eur Rev Med Pharmacol Sci 2020;24:8812-21. [PubMed]
- Wang D, Cai L, Tian X, et al. MiR-543 promotes tumorigenesis and angiogenesis in non-small cell lung cancer via modulating metastasis associated protein 1. Mol Med 2020;26:44. [Crossref] [PubMed]
- Fok KL, Chung CM, Yi SQ, et al. STK31 maintains the undifferentiated state of colon cancer cells. Carcinogenesis 2012;33:2044-53. [Crossref] [PubMed]
- Xiong J, Xing S, Dong Z, et al. STK31 regulates the proliferation and cell cycle of lung cancer cells via the Wnt/betacatenin pathway and feedback regulation by cmyc. Oncol Rep 2020;43:395-404. [PubMed]
- Watany MM, Elmashad NM, Badawi R, et al. Serum FBLN1 and STK31 as biomarkers of colorectal cancer and their ability to noninvasively differentiate colorectal cancer from benign polyps. Clin Chim Acta 2018;483:151-5. [Crossref] [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96-118. [Crossref] [PubMed]
- Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. [Crossref] [PubMed]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. [Crossref] [PubMed]
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer 2021;21:22-36. [Crossref] [PubMed]
- Safarpour-Dehkordi M, Doosti A, Jami MS. Integrative Analysis of lncRNAs in Kidney Cancer to Discover A New lncRNA (LINC00847) as A Therapeutic Target for Staphylococcal Enterotoxin tst Gene. Cell J 2020;22:101-9. [PubMed]
- Tu LR, Li W, Liu J, et al. LncRNA LINC00847 contributes to hepatocellular carcinoma progression by acting as a sponge of miR-99a to induce E2F2 expression. J Biol Regul Homeost Agents 2020;34:2195-203. [PubMed]
- Li H, Chen YK, Wan Q, et al. Long Non-coding RNA LINC00847 Induced by E2F1 Accelerates Non-small Cell Lung Cancer Progression Through Targeting miR-147a/IFITM1 Axis. Front Med (Lausanne) 2021;8:663558. [Crossref] [PubMed]
- Rao M, Xu S, Zhang Y, et al. Long non-coding RNA ZFAS1 promotes pancreatic cancer proliferation and metastasis by sponging miR-497-5p to regulate HMGA2 expression. Cell Death Dis 2021;12:859. [Crossref] [PubMed]
- Yang Y, Wang Y, Liu S, et al. How hsa-miR-495 performed in the tumorigenesis of pancreatic adenocarcinoma by bioinformatics analysis. J Cell Biochem 2018; Epub ahead of print. [Crossref] [PubMed]
- Yuan W, Gao H, Wang G, et al. Higher miR-543 levels correlate with lower STK31 expression and longer pancreatic cancer survival. Cancer Med 2020;9:9632-40. [Crossref] [PubMed]
- Kim H, Lee S, Shin E, et al. The Emerging Roles of Exosomes as EMT Regulators in Cancer. Cells 2020;9:861. [Crossref] [PubMed]
- The Lancet Gastroenterology H. Pancreatic cancer: a state of emergency? Lancet Gastroenterol Hepatol 2021;6:81. [Crossref]
- Tao J, Yang G, Zhou W, et al. Targeting hypoxic tumor microenvironment in pancreatic cancer. J Hematol Oncol 2021;14:14. [Crossref] [PubMed]
- Wang YC, He WY, Dong CH, et al. lncRNA HCG11 regulates cell progression by targeting miR-543 and regulating AKT/mTOR pathway in prostate cancer. Cell Biol Int 2019; Epub ahead of print. [Crossref] [PubMed]
- Wang H, Huang Y, Yang Y. LncRNA PVT1 Regulates TRPS1 Expression in Breast Cancer by Sponging miR-543. Cancer Manag Res 2020;12:7993-8004. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


