
PRRX1 promotes colorectal cancer stemness and chemoresistance via the JAK2/STAT3 axis by targeting IL-6
Highlight box
Key findings
• Overexpression of PRRX1 in CRC results in an enrichment of various gene sets, which played important roles in the development and progression of CRC. PRRX1 plays an important role in promoting the proliferation, viability and stemness of CRC cells through JAK2/STAT3 signaling by targeting IL6.
What is known and what is new?
• PRRX1 is a well-known stem cell-like properties regulator, and plays an important role in the development and progression of cancers. One study revealed a novel effect of PRRX1 on stemness acquirement of CRC cells, but its underlying mechanism remains unknown;
• PRRX1 was extensively increased in CRC, and tightly associated with the chemoresistance and poor prognosis of CRC. Silencing the expression of PRRX1 dramatically reduced the chemoresistance and metastasis rate of CRC
What is the implication, and what should change now?
• PRRX1 could be a valid biomarker for predicting the chemoresistance and poor prognosis of CRC, as well as being an important target for CRC therapeutic management.
Introduction
Colorectal cancer (CRC) is the third most common type of cancer in men and the second most common in women. The International Agency for Research on Cancer declared that 1.8 million new cases of CRC were diagnosed worldwide in 2018 (1). Owing to the contribution of early screening, the morbidity and mortality rates of CRC have been declining in some developed regions (2); however, those rates are still extremely high, especially among individuals with a Black racial/ethnic background (3). CRC is the second leading cause of cancer death globally and was estimated to account for 0.86 million deaths in 2018 (1). Tumor metastasis and chemoresistance are the 2 major factors of a poor prognosis of CRC patients, but their underlying molecular mechanisms are still not clear (4-6).
CRC is composed of heterogeneous cell populations, including those characterized with self-renewal and multi-lineage differentiation, which have been termed cancer stem cells (CSCs) (7). Accumulating evidence suggests that the acquisition of stemness in cancer cells significantly promote the recurrent metastasis and chemoresistance of CRC, leading to the mortality of CRC patients (8). It is well known that CSCs are characterized with ectopic high expression of multi-stemness factors, and can be identified with specific markers such as CD44, CD133, and ALDH1 (7). However, where and how the CSCs originate remains controversial. Some studies propose that CRC cells acquire stemness via dedifferentiation, yet others suggest that they are transformed from intestinal stem cells (7,9). Considering the important role of cell stemness in the development, progression, and prognosis of tumors, killing CSCs and/or inhibiting the acquisition of cell stemness could be a critical component for effective antitumor therapy in CRC patients.
Paired related homeobox 1 (PRRX1, also known as PRX1) is a transcriptional factor expressed during early limb bud mesoderm development (10). Studies have demonstrated that PRRX1 is an epithelial-to-mesenchymal transition (EMT) inducer, as well as a stem cell-like properties regulator, and plays a vital role in the development and progression of cancers (9,11-15). Previous studies on CRC have also revealed that PRRX1 plays a vital role in tumor metastasis by promoting EMT (13,16,17). It is understood that the acquisition of stemness is significantly related to the chemoresistance of cancers, which is a major contributor to a poor prognosis (18,19). However, the role of PRRX1 in regulating the stemness of CRC cells and chemoresistance is still not clear.
Interleukin 6 (IL-6) is a well-known cytokine, involved in the development and progression of cancers, including CRC (20). Study in mice has confirmed that blocking IL-6 can significantly reduce the colitis-associated tumorigenesis (21). In addition, studies have also revealed that IL-6 can promote the proliferation of CRC cells by activating STAT, ERK/MAPK and PI3K/ATK pathway (22). Moreover, recent study revealed that increase expression of IL-6 significantly promote the resistance of CRC cells to chemotherapy (23).
The present study aimed to uncover the role of PRRX1 in the chemoresistance of CRC and gain further insight into the importance of PRRX1/IL-6 axis in increasing the acquisition of stem cell-like properties reduces drug sensitivity. We present the following article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1137/rc).
Methods
Clinical samples
The fresh primary CRC and paired normal colorectal samples (64 cases), as well as the paraffin samples (101 cases of tumor tissues from CRC patients and 46 cases of normal colorectal tissue from healthy patients), were obtain from tissue bank of the Zhujiang Hospital, Southern Medical University, Guangzhou, China, from 2007 to 2018. The distances between cancer and paired adjacent normal tissues measured at least 2 cm. The pathological diagnosis of each case was made by the Department of Pathology in Zhujiang Hospital. We restricted our analysis to sporadic cases and did not make selections on the basis of family history. This study was approved by the Ethics Committees of Zhujiang Hospital, Southern Medical University, and comply with the criteria established by the Declaration of Helsinki (as revised in 2013). Individual consent for this retrospective analysis was waived. Target gene expression profiling studies of CRC samples, including relevant clinical information, were identified through searching in the Gene Expression Omnibus (GEO): GSE18088 (n=53), GSE87211 (n=363), and GSE39582 (n=585). The final expression level of PRRX1 in fresh samples from our hospital, as well as samples from GEO datasets GSE18088, GSE87211, and GSE39582, were defined as low (less than median value) or high (greater than median value).
Cell culture and treatment
The colorectal normal epithelial cell line FHC (Cat# CRL-1831) and the cancer cell lines HT29 (Cat# HCT-38), HCT116 (Cat# CCL-247), LS174T (Cat# CL-188), SW480 (Cat# CCL-228), SW620 (Cat# CCL-227), and RKO (Cat# CRL-2577) were obtained from Foleibao Biotechnology Development Company (Shanghai, China). For use in this study, the cells were propagated for less than 6 months after resuscitation. All cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium (Cat# A1049101; Thermo Fisher Life Technologies Corporation, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Cat# 10091130; Invitrogen, Waltham, MA, USA). A lentivirus with PRRX1 oligonucleotides or PRRX1 short hairpin RNA (shRNA) was purchased from GeneCopoeia (Guangzhou, China) and transfected as previously described (24). A vector with interleukin-6 (IL-6) shRNA (sh-IL-6) was purchased from GeneChem (Shanghai, China) and transfected using lipofectamine® 3000 reagent (Cat# L3000015; Thermo Fisher Scientific, USA). The duration of chemotherapy treatment in the in vitro assay was 24 hours.
RNA isolation, reverse transcription (RT), and quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted using Trizol (Invitrogen; Carlsbad, CA, USA). To quantify the expression of PRRX1, IL-6, and IL-6 cytokine family signal transducer (IL6ST), we subjected the total RNA to polyadenylation and RT using a ThermoScriptTM RT-PCR System (Invitrogen). RT-PCR analysis was carried out using an SYBR Green PCR master mix (Applied Biosystems; Foster City, CA, USA) on an ABI 7500HT system. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) small nuclear RNA (snRNA) was used as an endogenous control. All samples were normalized to internal controls, and fold changes were calculated through relative quantification (2−ΔΔCt). The RT-PCR for target genes was performed as previously described (25). The primers used are shown in Table S1. The subject was classified as the PRRX1-high expression group if its PRRX1 expression was higher than the average value, otherwise it was classified as the PRRX1-low expression group.
Immunoblotting assay
For immunoblotting, cultured cells were harvested, washed with phosphate-buffered saline (PBS), and lysed in radioimmunoprecipitation assay (RIPA) buffer as described previously (24). Antibodies used are listed as follows: rabbit antibodies against PRRX1 (Cat# ab211292) and Nanog (Cat# ab218524) were from Abcam (Shanghai, China). Rabbit antibodies against CD133 (Cat# 64326), SOX2 (Cat# 3579), OCT4 (Cat# 2750), phosphorylated-JAK2 (Cat# 3771), phosphorylation-STAT3 (Cat# 9145), and cleaved-PARP (Cat# 5625) were from Cell Signaling Technology (CST; Danvers, MA, USA). Rabbit antibodies against JAK2 (Cat# 17670-1-AP), mouse against IL-6 (Cat#66146-1-Ig), STAT3 (Cat# 60199-1-Ig), BCL2 (Cat# 60178-1-Ig), CNND1 (Cat# 60186-1-Ig), and PARP (Cat# 66520-1-Ig) and GAPDH (Cat# 60004-1-Ig) were from Proteintech (Rosemont, IL, USA). All the antibodies were used at 1:1,000 dilutions. The membranes, probed with the indicated primary antibodies, were subjected to the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (anti-rabbit: Cat# 7074 and anti-mouse: Cat# 7076, CST, USA), and developed by enhanced chemiluminescence.
Immunohistochemistry (IHC) assay
IHC was performed on paraffin sections of normal colorectal and CRC tissues according to standard labelled streptavidin-biotin (LSAB) protocol (Dako) using primary antibodies against PRRX1 (Cat# ab211292, Abcam, China). The degree of staining in the sections was observed and scored independently by two pathologists. The percent positivity of PRRX1 staining was scored from 0 to 4: 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (>75%). The staining intensity was scored on a 4-point scale: 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellowish-brown), and 3 (strong staining, brown). Subsequently, the PRRX1 expression score was calculated as the product of the percent positivity score and staining intensity score and ranged from 0 to 12. The final expression level of PRRX1 was defined as low [0–5] or high [6–12].
Immunofluorescence assay
Cells were cultured on coverslips overnight, fixed with 4% paraformaldehyde for 30 minutes, and then treated with 0.5% Triton X-100 for 15 minutes. After being blocked in 10% normal blocking serum at room temperature for 15 minutes, the slides were incubated with antibodies from CST (USA), including rabbit antibodies to PRRX1 (Cat# DF4274; Affinity, Cincinnati, OH, USA), CD133 (Cat# 64326; CST, USA) and SOX2 (Cat# 3579; CST, USA) (1:100) at 4 ℃ overnight, which was followed by PBS rinsing. Cover slips were incubated with antibodies from CST (USA), including a fluorescein isothiocyanate (FITC)-conjugated anti-rabbit (Cat# 4412) or mouse (Cat# 4408) stain and Texas Red-conjugated anti-mouse (Cat# 8890) or anti-rabbit antibodies (Cat# 8889) (1:200) for 30 minutes at room temperature, then stained with 6-diamidino-2-phenylindole (DAPI; Cat# D3571, Invitrogen, USA).
Enzyme-linked immunosorbent assay (ELISA)
The supernatants and cytoplasm from SW480 and HCT116 cells were collected. The supernatant was used to measure the total levels of extracellular IL-6 by the human IL-6 ELISA kit (Cat# ab178013; Abcam, USA), according to the manufacturer’s introductions. The cytokine expression level (pg/mL) per 105 cells was analyzed.
Cell apoptosis analysis
Cells (5×105) were washed with ice-cold PBS then resuspended in 500 µL of binding buffer (KeyGen Biotechnology, Nanjing, China). Annexin V-FITC (5 µL) (KeyGen) and propidium iodide (PI; 5 µL) were added to the cell suspension. The mixture was then incubated at room temperature for 10 minutes in the dark. Analysis of apoptosis was carried out using flow cytometry.
Cell proliferation and colony formation assays
Cell proliferation assays were carried out using a Cell Counting Kit-8 (CCK-8; Dojindo; Kumamoto, Japan). Cells were plated in 96-well plates at a density of 1×104 cells per well and were cultured in the growth medium. At the indicated time points, the number of cells in triplicate wells was measured at an absorbance at 450 nm of reduced WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfo-phenyl)-2H-tetrazolium monosodium salt].
For the colony-formation assay, 500 viable cells were placed in each well in 6-well plates and maintained in a complete medium for 2 weeks. Colonies were fixed with methanol and stained with 0.1% (w/v) crystal violet.
Cell-cycle analysis and 5-ethynyl-2'-deoxyuridine incorporation assay
For cell-cycle analysis, a total number of 5×106 cells were harvested and then washed with cold PBS. The cells were further fixed with 70% ice-cold ethanol at 4 ℃ overnight. After incubation with PBS containing 10 mg/mL PI and 0.5 mg/mL RNase A for 15 minutes at 37 ℃, fixed cells were washed with cold PBS 3 times. We used FACScaliber flow cytometry (Becton, Dickinson, and Co. BD Biosciences, Franklin Lakes, NJ, USA) was used to gain the DNA content of labeled cells.
For the 5-ethynyl-2'-deoxyuridine (EdU) incorporation assay, proliferating cells were examined using the Keyfluor488-EdU Cell Proliferation Assay Kit (KeyGen Biotechnology, Nanjing, China) according to the manufacturer’s protocol. Briefly, after incubation with 50 µM EdU for 2 hours, cells were fixed with 4% paraformaldehyde, permeabilized in 0.5% Triton X-100, and stained with Apollo fluorescent dyes. A total of 5 µg/mL of DAPI was used to stain cell nuclei for 15 minutes. The number of EdU-positive cells was counted under a fluorescent microscope in 5 random fields (original magnification: ×200). All assays were independently performed 3 times.
Sphere-forming assay
Suspensions of single-cells were seeded into 6-well plates at a density of 5,000 cells/mL in stem cell-conditioned medium containing Dulbecco’s modified Eagle medium (DMEM)/F12 supplemented with 100 IU/mL penicillin G, 100 µg/mL streptomycin, 10 ng/mL epidermal growth factor (EGF; Peprotech Inc., Rocky Hill, NJ, USA), 10 ng/mL basic fibroblast growth factor (bFGF; Peprotech Inc., USA), and 1× B27 (Invitrogen, USA). The sphere-forming cells were analyzed by counting the number of cells and spheres on day 7. Culture suspensions were not passaged during the whole experiment.
Luciferase reporter assays
Using JASPAR software (https://jaspar.genereg.net/), IL-6 was predicted to be the target gene of PRRX1. A 2,000-bp fragment containing 2 binding sites of IL-6 promoter [named wild type (WT)] was PCR-amplified and inserted into a psiCHECK-2 luciferase reporter vector. In addition, IL-6-binding sites mutations [Mutation-1: Δ(585–592), Mutation-2: Δ(1,088–1,095)] were constructed. All the psiCHECK-2 vectors were co-transfected with PRRX1-overexpression vector into 293T cells using the lipofectamine® 3000 reagent. Luciferase activity was measured at 48 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega Corp., Madison, WI, USA).
PRRX1 binding motifs and potential binding site of target gene analysis
The binding motifs of PRRX1 and its potential binding sites on target genes were predicted by the public bioinformational analysis software JASPAR2020. Some 2,000 base pairs prior to the coding sequence of the target genes were chosen as the prediction region for the promoter binding site. The relative profile score threshold was 95%. Only the sites on the sense strands were considered potential binding sites in our study.
Orthotopic xenograft CRC mouse model
HCT116 cells (transfected with control or PRRX1-shRNA lentivirus) were suspended in fresh PBS at a concentration of 1×106 cells/50 µL and aspirated using a fine needle. Then, 6-week-old BALB/c nude mice (male, about 30-gram weight) were anaesthetized, and the cecum of each was exposed by laparotomy. In brief, a 0.5–1 cm-long nick was made in the skin, and the abdominal wall musculature was lifted. The abdominal cavity was opened, and the cecum was isolated and covered by sterile gauze and warm saline to keep moist. The HCT116 cells (50 µL) were slowly injected into the cecal wall. The needle was carefully removed, and the injection site was inspected to ensure no leakage. The cecum was then put back into the abdominal cavity and the abdominal wall and skin were sutured. A week after cell injections, either 30 mg/kg 5-fluorouracil (5-FU) (Cat# F5130; Sigma, St. Louis, MO, USA) or 5 mg/kg oxaliplatin (L-OHP) (Cat# O9512; Sigma, USA) was injected intraperitoneally to the corresponding groups twice a week. In vivo imaging was performed at days 7, 16, and 28 after cell injection, with luciferin (150 mg/kg) (Cat# P1043; Caliper Life Science, Hopkinton, MA, USA) intraperitoneally injected into each mouse 10 minutes before imaging. Caliper IVIS Lumina II (Caliper Life Sciences, USA) was used for bioluminescence imaging to monitor tumor growth. After 28 days, the mice were sacrificed, and the orthotopic xenograft CRC masses were measured and harvested for further hematoxylin and eosin (HE) study. A protocol was prepared before the study without registration. Experiments were approved by the Ethics Committee of Zhujiang Hospital, Southern Medical University, in compliance with institutional guidelines for the care and use of animals.
Statistical analysis
The data were analyzed using the software SPSS 19.0 (IBM Corp., Chicago, IL, USA). The clinical data were analyzed using nonparametric tests (Wilcoxon and Mann-Whitney) as well as Kaplan-Meier and Cox regression survival analyses. Pearson’s chi-squared (χ2) test, unpaired Student’s t-test, and paired t-test were used to evaluate the significance of the differences among different groups. All statistical tests were two-sided. The data are presented as the means ± standard error of the means (SEMs).
Results
High expression of PRRX1 was closely associated with tumor metastasis, chemoresistance, and poor prognosis of CRC patients
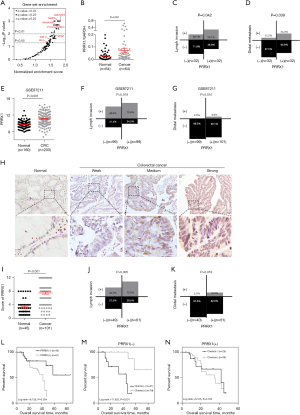
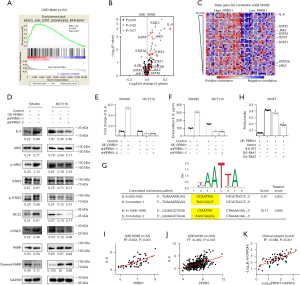
Gene set enrichment analysis (GSEA) of 53 CRC tissue samples from the GEO dataset GSE18088 showed that various signaling pathways, which are closely associated with the metastasis and stemness of CRC, were enriched in the PRRX1-high expression group, including the JAK/STAT (Janus kinase/signal transducer and activator of transcription), focal adhesion, Wnt, and transforming growth factor-β (TGF-β) signaling pathways, indicating the crucial role of PRRX1 in the proliferation, metastasis, and stemness properties of CRC cells (Figure 1A, Table S1). We further detected the transcriptional level of PPRX1 in 64 pairs of CRC tissues and matched normal mucosa, finding that PRRX1 was overexpressed in CRC tissues and closely related to a high risk of lymph and distal metastasis (Figure 1B-1D, Table S2). Consistent results were also found in samples from the GEO GSE87211 dataset (Figure 1E-1G). Further investigation of the expression of PRRX1 in 101 CRC and 46 normal control samples also revealed an increased expression of PRRX1 in tumor tissues, and a positive correlation between PRRX1 expression and the incidence of tumor metastasis (Figure 1H-1K, Table S3). Moreover, high expression of PRRX1 in tumor tissues indicated a poor prognosis for CRC patients (Figure 1L). Notably, chemotherapy could dramatically improve the prognosis of CRC patients characterized with low expression of PRRX1 (Figure 1M,1N), indicating an important role of PRRX1 in enhancing the chemoresistance of CRC cells. Consistent conclusions were reached while analyzing the GEO GSE103479 dataset (Figure S1). Collectively, this indicated that PRRX1 might exert great effects on CRC through regulating the metastasis and chemoresistance of tumors.
PRRX1 promoted the proliferation and stemness properties of CRC cells in vitro and in vivo
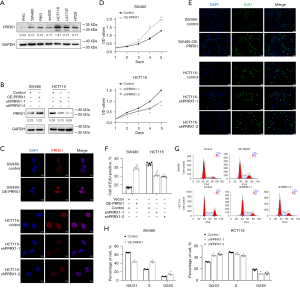
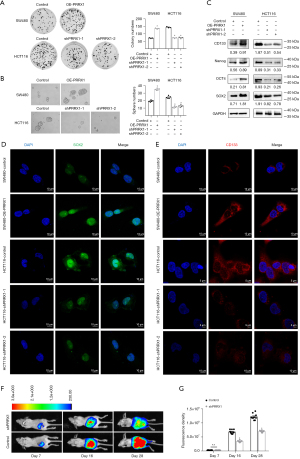
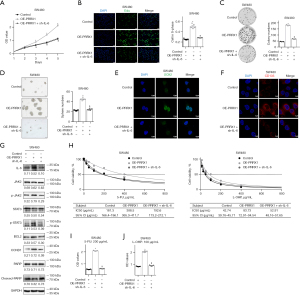
The transcriptional levels of PRRX1 were detected in 6 CRC cell lines (SW480, RKO, SW620, HCT116, LS174T, and HT29) and normal epithelial cells (FHC), and we chose SW480 and HCT116 for further analysis (Figure 2A). Next, the expression of PRRX1 was overexpressed in SW480 cells and silenced in HTC116 cells by using lentivirus with PRRX1 oligonucleotides (OE-PRRX1) and shRNA (shPRRX1), respectively (Figure 2B, Figure S2). Additional immunofluorescence assays showed a consistent alteration of PRRX1 expression in both SW480 and HCT116 cells, especially in their cell nuclei (Figure 2C). The CCK-8 assay showed that overexpressing PRRX1 significantly promoted the proliferation of SW480 cells, whereas decreasing PRRX1 had an opposite effect in HCT116 cells (Figure 2D). Further EdU incorporation (Figure 2E,2F) and flow cytometry (Figure 2G,2H) assays revealed that an increased expression of PRRX1 dramatically increased the percentage of SW480 cells in the S phase of the cell cycle, whereas silencing PRRX1 decreased that percentage in HCT116 cells, which also indicated the promotion effect of PRRX1 on cell proliferation. In addition, we found that overexpressing PRRX1 remarkably enhanced the colony (Figure 3A) and sphere (Figure 3B) formation properties of CRC cells in SW480 cells, whereas decreasing PRRX1 significantly inhibited these 2 properties in HCT116 cells. Further Western blot assay (Figure 3C) and immunofluorescence staining (Figure 3D,3E) found a consistent alteration of stemness signature in both SW480 and HCT116 cells. We also found that decreasing the expression of PRRX1 in HCT116 cells significantly inhibited the proliferation of CRC in vivo (Figure 3F,3G). Taken together, these results revealed that PRRX1 had a great effect on promoting the proliferation and stemness of CRC cells.
PRRX1 enhanced the resistance of CRC cells to 5-FU and L-OHP in vitro
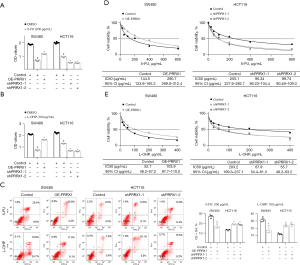
Since high expression of PRRX1 is closely associated with a poor response to chemotherapy in CRC patients, we investigated the role of PRRX1 in enhancing the resistance of CRC cells to both 5-FU and L-OHP. The CCK-8 assay showed that an increased expression of PRRX1 significantly enhanced the viability of SW480 cells after treatment with 5-FU or L-OHP, whereas decreasing the expression of PRRX1 achieved the opposite results (Figure 4A,4B). Flow cytometry also found that PRRX1 dramatically decreased the percentage of CRC cells undergoing apoptosis during treatment with 5-FU or L-OHP (Figure 4C). Furthermore, half maximal inhibitory concentration (IC50) assays showed that overexpressing PRRX1 remarkably reduced the sensitivity of SW480 cells to both 5-FU and L-OHP, whereas silencing PRRX1 enhanced their sensitivity in HCT116 cells (Figure 4D,4E).
Down-regulation of PRRX1 enhanced the sensitivity of CRC cells to 5-FU and L-OHP in vivo
An orthotopic xenograft CRC mouse model was applied to further investigate the effect of PRRX1 on chemoresistance in vivo (Figure 5A). In vivo imaging assays showed that knocking down the expression of PRRX1 significantly enhanced the sensitivity of HCT116 cells to both 5-FU and L-OHP (Figure 5B-5D). Silencing PRRX1 reduced the liver metastasis rate of mice from 60% to 10% in 5-FU treatment groups, and from 70% to 10% in L-OHP treatment groups (Figure 5E-5G). However, no significant difference of mice body weight was found between the 2 groups in both 5-FU and L-OHP treatment (Figure S3). Accordingly, we considered that PPRX1 might play a vital role in promoting the chemoresistance of CRC cells.

PRRX1 activated the JAK2/STAT3 axis by targeting IL-6
The GSEA revealed that a high expression of PRRX1 in CRC was closely associated with the activation of multiple cancer-related signaling pathways, among which JAK/STAT signaling had the highest normalized enrichment score (Figure 6A, Table S1). Further analysis of data from the GEO GSE18088 dataset suggested that PRRX1 significantly increased both the transcriptional level (Figure 6B) and the ranking in the gene list (Figure 6C, Table S4) of IL-6, an important trigger for the activation of JAK2/STAT3 signaling (26). It is well known that the JAK2/STAT3 signaling plays an important role in enhancing the stemness of cancer cells (26,27). Thus, we suspected that PRRX1 might promote the stemness and chemoresistance of CRC cells via the IL-6/JAK2/STAT3 axis. Further study demonstrated that the expression of IL-6 significantly increased in CRC tissues when compared with adjacent normal tissues (Figure S4). PRRX1 dramatically increased the transcription, intracellular expression, and extracellular expression of IL-6, as well as the expression of phosphorylated JAK2 and STAT3. Moreover, PRRX1 significantly promoted the expression of BCL2 and CCND1, downstream of IL-6/JAK2/STAT3, and it also decreased the expression of cleaved-PARP (Figure 6D-6F). These findings strongly indicated the activation of the IL-6/JAK2/STAT3 axis. The publicly available bioinformatics algorithm JASPAR suggested that IL-6 was one target gene of PRRX1 (Figure 6G). Further luciferase assays confirmed that PRRX1 had bound to the promoter of IL-6 at both Δ(585–592) and Δ(1,088–1,095) sites away from the 5' terminal of coding sequence, but the Δ(1,088–1,095) site was the primary regulating domain of PRRX1 on IL-6 transcription (Figure 6H). Moreover, high correlations between PRRX1 and IL-6 expression were also found in the clinical CRC samples from our hospital as well as GEO GSE18088 and GSE39582 datasets (Figure 6I-6K).
PRRX1 enhanced stemness and chemoresistance of CRC cells via the IL-6/JAK2/STAT3 axis
To clarify the role of the IL-6/JAK2/STAT3 axis in PRRX1-mediated stemness and chemoresistance of CRC cells, we disturbed the expression of IL-6 in a SW480 cell line with highly expressed PRRX1 by using a vector with shRNA targeting IL-6 (Figure S5), and verified its role in regulating the activation of JAK/STAT signaling, cell proliferation, and stemness, as well as sensitivity to chemotherapy (Figure S6). The CCK-8 and EdU incorporation assays showed that disturbing the expression of IL-6 significantly eliminated the promotion of PRRX1 in CRC cell proliferation (Figure 7A,7B). Further study also revealed that silencing IL-6 dramatically reduced the enhancement of PRRX1 in colony and sphere formation of CRC cells (Figure 7C,7D). Similarly, disturbing IL-6 expression also significantly reversed the PRRX1-mediated alteration of stemness signatures (SOX2, CD133) and JAK2/STAT3 signaling (p-JAK2, p-STAT3, BCL2, CCND1, and cleaved-PARP) (Figure 7E-7G). Silencing IL-6 also significantly eliminated the promotion of PRRX1 in IC50 and resistance of CRC to both 5-FU and L-OHP (Figure 7H-7J). In accord with the above findings, we concluded that PRRX1 promoted the stemness and chemoresistance of CRC cells primarily via the IL-6/JAK2/STAT3 axis.
Discussion
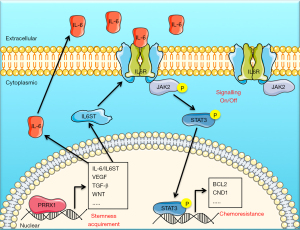
Studies have shown that PRRX1 plays a vital role in promoting the migration/invasion of cells, and it has also been speculated to be a valid biomarker for predicting the risk of tumor metastasis of various cancers, including CRC (13,16,28), liver cancer (29), breast cancer (30), and pancreatic cancer (31,32). The present study highlights the important role of PRRX1 in chemotherapy treatment of CRC, as we found that chemotherapy could improve the prognosis of only those CRC patients with low PRRX1 expression. Moreover, our bioinformatics analysis of clinical CRC samples also showed that a high expression of PRRX1 not only significantly promoted the enrichment of gene sets involved in cell migration/invasion, but also promoted gene sets involved in cell proliferation, viability, and stemness, which play important roles in enhancing the resistance of cancer cells to anti-cancer drugs. Further in vitro and in vivo investigations demonstrated the regulation of PRRX1 on CRC cell chemoresistance. PRRX1 dramatically promoted the proliferation, viability, and stemness properties of CRC through the JAK2/STAT3 axis by targeting IL-6. Thus, we theorized that PRRX1 could be a valid biomarker for predicting the sensitivity of CRC patients to chemotherapy, as well as a potential target for CRC chemotherapeutic management.
Since the activation of JAK/STAT signaling is closely related to the stemness acquisition of cancer cells (26,27), and this signaling had the highest enrichment score in the PRRX1-high expression group, it was considered the primary potential drive pathway for PRRX1-mediated stemness acquisition and chemoresistance of CRC in our study. Further analysis showed that high expression of PRRX1 dramatically promoted the enrichment of IL-6 in CRC cells, which is a well-known trigger for the activation of JAK/STAT signaling, especially the JAK2/STAT3 axis (26). Thus, we firstly explored the role of the IL-6/JAK2/STAT3 axis in PRRX1-mediated stemness and chemoresistance of CRC. Intriguingly, we found a direct promotion of PRRX1 in IL-6 on an intracellular and extracellular level, and a consistent alteration to it downstream. We also found a high correlation between the transcriptional expressions of PRRX1 and IL-6 in clinical tumor samples. These findings strongly indicated that PRRX1 was a major transcriptional factor for regulating the transcription of IL-6 in CRC. In addition, we found that the promotions of PRRX1 on stemness and chemoresistance of CRC cells, as well as related signatures, were significantly eliminated by disturbing the expression of IL-6. Thus, we concluded that PRRX1 promoted the stemness and chemoresistance of CRC primarily via the IL-6/JAK2/STAT3 axis.
We also found that a high expression of PRRX1 significantly increased the rank list of other elements of the JAK/STAT signaling pathway in cells, including JAK3, STAT2, STAT4, and STAT5A (33). However, only JAK3, STAT2, and STAT4 showed high correlations with PRRX1 in clinical CRC samples. We suspected that the ranking change of STAT5A in the gene list might be a secondary change following the change of other genes, which accounts for the ranking change of JAK2 in our study.
Many studies have demonstrated the critical role of JAK3 in the proliferation and differentiation of lymph cells (34,35). However, its effect on CRC cell bioactivity is still not clear. As for STAT2, it is specifically activated by type I interferons-α/β (IFN-α/β) and is mainly involved in regulating the proliferation and differentiation of lymph cells (36). A recent study revealed that STAT2 coordinates with interferon regulatory factor 9 (IRF9) and promotes the expression of IL-6 (37). Therefore, it is conceivable that PRRX1 might indirectly promote the expression of IL-6 by targeting STAT2 in CRC cells. As for STAT4, a study revealed its effect on promoting the proliferation and migration/invasion of cancer cells (38,39), meaning that PRRX1 may also promote the development and metastasis of CRC by directly targeting STAT4. Although our study focused on revealing the vital role of the IL-6/JAK2/STAT3 axis in PRRX1-mediated stemness acquisition and chemoresistance of CRC, these results imply that PRRX1 may also promote the development and progression of CRC by targeting other elements of JAK/STAT signaling.
Our study also found that PRRX1 significantly promoted the transcription of gene IL-6 signaling transducer (IL6ST, also known as GP130 or CD130). A high correlation of its transcriptional expression was found in clinical CRC samples. Moreover, bioinformatics analysis by JASPAR software showed that the promoter of IL6ST also had a binding site for PRRX1 (Supplementary Figure S7). These findings strongly suggested that PRRX1 might also be a major transcriptional regulator for IL6ST. Additionally, it is widely known that IL6ST plays an important role in transducing the signaling of IL-6 from the cell membrane to cytoplasm (40). It is conceivable that a high expression of IL6ST could more completely promote the activation of the JAK2/STAT3 axis and further enhance the stemness and chemoresistance of CRC cells. Therefore, even without direct evidence, we speculated that PRRX1 might sufficiently promote the activation of the IL-6/JAK2/STAT3 axis by targeting IL6ST.
Studies have demonstrated that various biological factors, including Snail (41), SIRT1 (9), and HSP27 (42), have great effects on regulating the expression of PRRX1 in cancers. As for CRC, 2 different micro-RNAs (miRNA), miR-106b and miR-124, have been identified as important upstream regulators of PRRX1 (16,28). These findings strongly suggested that PRRX1 might be a common downstream product of various etiological factors during the development and progression of cancers and could be a potential target for CRC therapeutic management. However, considering the complex upstream network of PRRX1, as well as its extensive regulation of cell signaling, it was difficult to eliminate the entire effect of PRRX1 on CRC cells and not disturb the expression and/or nuclear localization of PRRX1 itself.
Since PRRX1 is a well-known inducer of EMT (12), many studies consider PRRX1 to promote tumor metastasis mainly by inducing EMT in cancer cells (9,13). However, our analysis found that PRRX1 also dramatically promoted the enrichment of gene set “focal adhesion”, the activation of which is also a crucial risk factor for tumor metastasis, signifying that the alteration of focal adhesion might be another important mechanism underlying PRRX1-mediated CRC metastasis. It was also conceivable that PRRX1 might play an important role in other cell activities, because high expression of PRRX1 significantly promoted the enrichment of various key cancer-related gene sets, including mitogen-activated protein kinase (MAPK) (43), TGF-β (44), and vascular endothelial growth factor (VEGF) (45). Each of these signaling pathways plays a critical role in the response and inhibition of immunity (43-45), indicating that a high expression of PRRX1 in CRC patients might be an important etiological factor contributing to a poor prognosis for those patients after treatment with immunotherapy. Ultimately, further investigation on the role of PRRX1 in the development and progression of CRC is still necessary.
Conclusions
The present study highlights the effects of PRRX1 on stemness and chemoresistance of CRC through the IL-6/JAK2/STAT3 axis (Figure 8). PRRX1 could be a valid biomarker for predicting the sensitivity of CRC patients to chemotherapy, as well as a potential target for CRC therapeutic management. Further investigation of the role of PRRX1 in the development and progression of CRC is still necessary.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Guangdong Province (Nos. 2020A1515010212 and 2019A1515010922); the Science and Technology Program of Guangzhou (No. 202102020903); the Medical Scientific Research Foundation of Guangdong Province of China (No. B2018002); the Natural Science Foundation of China (Nos. 81901987 and 82103399).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1137/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1137/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1137/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- International Agency for Research on Cancer, Cancer Incidence and Mortality Worldwide: IARC CancerBase, 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014;64:104-17. [Crossref] [PubMed]
- Smith RA, Andrews KS, Brooks D, et al. Cancer screening in the United States, 2018: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin 2018;68:297-316. [Crossref] [PubMed]
- Hu Z, Ding J, Ma Z, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet 2019;51:1113-22. [Crossref] [PubMed]
- Ebert MP, Tänzer M, Balluff B, et al. TFAP2E-DKK4 and chemoresistance in colorectal cancer. N Engl J Med 2012;366:44-53. [Crossref] [PubMed]
- Zheng K, Yu J, Chen Z, et al. Ethanol promotes alcohol-related colorectal cancer metastasis via the TGF-beta/RUNX3/Snail axis by inducing TGF-beta1 upregulation and RUNX3 cytoplasmic mislocalization. Ebiomedicine 2019;50:224-37. [Crossref] [PubMed]
- Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer 2014;14:468-80. [Crossref] [PubMed]
- Medema JP. Targeting the Colorectal Cancer Stem Cell. N Engl J Med 2017;377:888-90. [Crossref] [PubMed]
- Shi L, Tang X, Qian M, et al. A SIRT1-centered circuitry regulates breast cancer stemness and metastasis. Oncogene 2018;37:6299-315. [Crossref] [PubMed]
- Logan M, Martin JF, Nagy A, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 2002;33:77-80. [Crossref] [PubMed]
- Greenbaum A, Hsu YM, Day RB, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 2013;495:227-30. [Crossref] [PubMed]
- Ocaña OH, Córcoles R, Fabra A, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 2012;22:709-24. [Crossref] [PubMed]
- Takahashi Y, Sawada G, Kurashige J, et al. Paired related homoeobox 1, a new EMT inducer, is involved in metastasis and poor prognosis in colorectal cancer. Br J Cancer 2013;109:307-11. [Crossref] [PubMed]
- Chen Z, Chen Y, Li Y, et al. Prrx1 promotes stemness and angiogenesis via activating TGF-β/smad pathway and upregulating proangiogenic factors in glioma. Cell Death Dis 2021;12:615. [Crossref] [PubMed]
- Lee KW, Yeo SY, Gong JR, et al. PRRX1 is a master transcription factor of stromal fibroblasts for myofibroblastic lineage progression. Nat Commun 2022;13:2793. [Crossref] [PubMed]
- Zhang Y, Zheng L, Huang J, et al. MiR-124 Radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One 2014;9:e93917. [Crossref] [PubMed]
- Du W, Liu X, Yang M, et al. The Regulatory Role of PRRX1 in Cancer Epithelial-Mesenchymal Transition. Onco Targets Ther 2021;14:4223-9. [Crossref] [PubMed]
- Cazet AS, Hui MN, Elsworth BL, et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat Commun 2018;9:2897. [Crossref] [PubMed]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer 2005;5:275-84. [Crossref] [PubMed]
- Zhang X, Hu F, Li G, et al. Human colorectal cancer-derived mesenchymal stem cells promote colorectal cancer progression through IL-6/JAK2/STAT3 signaling. Cell Death Dis 2018;9:25. [Crossref] [PubMed]
- Li ZW, Sun B, Gong T, et al. GNAI1 and GNAI3 Reduce Colitis-Associated Tumorigenesis in Mice by Blocking IL6 Signaling and Down-regulating Expression of GNAI2. Gastroenterology 2019;156:2297-312. [Crossref] [PubMed]
- Hsu CP, Chen YL, Huang CC, et al. Anti-interleukin-6 receptor antibody inhibits the progression in human colon carcinoma cells. Eur J Clin Invest 2011;41:277-84. [Crossref] [PubMed]
- Hu F, Song D, Yan Y, et al. IL-6 regulates autophagy and chemotherapy resistance by promoting BECN1 phosphorylation. Nat Commun 2021;12:3651. [Crossref] [PubMed]
- Zheng K, Zhou X, Yu J, et al. Epigenetic silencing of miR-490-3p promotes development of an aggressive colorectal cancer phenotype through activation of the Wnt/β-catenin signaling pathway. Cancer Lett 2016;376:178-87. [Crossref] [PubMed]
- Wang H, An H, Wang B, et al. miR-133a represses tumour growth and metastasis in colorectal cancer by targeting LIM and SH3 protein 1 and inhibiting the MAPK pathway. Eur J Cancer 2013;49:3924-35. [Crossref] [PubMed]
- Huang W, Zhong Z, Luo C, et al. The miR-26a/AP-2α/Nanog signaling axis mediates stem cell self-renewal and temozolomide resistance in glioma. Theranostics 2019;9:5497-516. [Crossref] [PubMed]
- Huang Y, Zhou B, Luo H, et al. ZnAs@SiO(2) nanoparticles as a potential anti-tumor drug for targeting stemness and epithelial-mesenchymal transition in hepatocellular carcinoma via SHP-1/JAK2/STAT3 signaling. Theranostics 2019;9:4391-408. [Crossref] [PubMed]
- Zheng L, Zhang Y, Lin S, et al. Down-regualtion of miR-106b induces epithelial-mesenchymal transition but suppresses metastatic colonization by targeting Prrx1 in colorectal cancer. Int J Clin Exp Pathol 2015;8:10534-44. [PubMed]
- Hirata H, Sugimachi K, Takahashi Y, et al. Downregulation of PRRX1 Confers Cancer Stem Cell-Like Properties and Predicts Poor Prognosis in Hepatocellular Carcinoma. Ann Surg Oncol 2015;22:S1402-9. [Crossref] [PubMed]
- Lv ZD, Kong B, Liu XP, et al. miR-655 suppresses epithelial-to-mesenchymal transition by targeting Prrx1 in triple-negative breast cancer. J Cell Mol Med 2016;20:864-73. [Crossref] [PubMed]
- Takano S, Reichert M, Bakir B, et al. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev 2016;30:233-47. [Crossref] [PubMed]
- Marchand B, Pitarresi JR, Reichert M, et al. PRRX1 isoforms cooperate with FOXM1 to regulate the DNA damage response in pancreatic cancer cells. Oncogene 2019;38:4325-39. [Crossref] [PubMed]
- Xin P, Xu X, Deng C, et al. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int Immunopharmacol 2020;80:106210. [Crossref] [PubMed]
- Nairismägi M. Oncogenic activation of JAK3-STAT signaling confers clinical sensitivity to PRN371, a novel selective and potent JAK3 inhibitor, in natural killer/T-cell lymphoma. Leukemia 2018;32:1147-56. [Crossref] [PubMed]
- Gigante M, Pontrelli P, Herr W, et al. miR-29b and miR-198 overexpression in CD8+ T cells of renal cell carcinoma patients down-modulates JAK3 and MCL-1 leading to immune dysfunction. J Transl Med 2016;14:84. [Crossref] [PubMed]
- Fan JB, Miyauchi S, Xu HZ, et al. Type I Interferon Regulates a Coordinated Gene Network to Enhance Cytotoxic T Cell-Mediated Tumor Killing. Cancer Discov 2020;10:382-93. [Crossref] [PubMed]
- Nan J, Wang Y, Yang J, et al. IRF9 and unphosphorylated STAT2 cooperate with NF-κB to drive IL6 expression. Proc Natl Acad Sci U S A 2018;115:3906-11. [Crossref] [PubMed]
- Zhao L, Ji G, Le X, et al. An integrated analysis identifies STAT4 as a key regulator of ovarian cancer metastasis. Oncogene 2017;36:3384-96. [Crossref] [PubMed]
- Cheng JM, Yao MR, Zhu Q, et al. Silencing of stat4 gene inhibits cell proliferation and invasion of colorectal cancer cells. J Biol Regul Homeost Agents 2015;29:85-92. [PubMed]
- Yonejima A, Mizukoshi E, Tamai T, et al. Characteristics of Impaired Dendritic Cell Function in Patients With Hepatitis B Virus Infection. Hepatology 2019;70:25-39. [Crossref] [PubMed]
- Fazilaty H, Rago L, Kass Youssef K, et al. A gene regulatory network to control EMT programs in development and disease. Nat Commun 2019;10:5115. [Crossref] [PubMed]
- Chen W, Ren X, Wu J, et al. HSP27 associates with epithelial-mesenchymal transition, stemness and radioresistance of salivary adenoid cystic carcinoma. J Cell Mol Med 2018;22:2283-98. [Crossref] [PubMed]
- Hugo W, Shi H, Sun L, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 2015;162:1271-85. [Crossref] [PubMed]
- Miao Y, Yang H, Levorse J, et al. Adaptive Immune Resistance Emerges from Tumor-Initiating Stem Cells. Cell 2019;177:1172-1186.e14. [Crossref] [PubMed]
- Apte RS, Chen DS, Ferrara N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019;176:1248-64. [Crossref] [PubMed]


