
Diagnostic genes and immune infiltration analysis of colorectal cancer determined by LASSO and SVM machine learning methods: a bioinformatics analysis
Introduction
Colorectal cancer has a relatively high incidence and mortality, with a gradually increasing incidence in recent years. According to statistics, it is the third most common cancer in the United States after breast cancer (prostate cancer for men) and lung cancer (1). Although advances in colonoscopy can provide early detection of colorectal cancer, and radiotherapy, chemotherapy, immunotherapy, and other treatment methods have improved the five-year survival rate to 65% (2), the annual number of deaths remains high at 52,980, accounting for 8.7% (52,980/608,570) (3) of all cancer deaths. Moreover, colorectal cancer in China is ranked among the top five diseases (4), and given that genetic factors account for approximately 35% of colorectal cancer risk (5) it is essential to study its pathogenesis further. The quantification of gastrointestinal tumor risk should be combined with clinical and molecular data to allow an accurate phenotypic assessment and genetic diagnosis (6). Several biomarkers have been identified recently for the diagnosis of colorectal cancer such as secretin receptor (SCTR) gene methylation (7), tRNA-derived small RNAs (tDRs) (8), long non-coding RNAs (lncRNAs) (9), and TMEM236 gene (10). However, the specificity and sensitivity of these diagnostic biomarkers for colorectal cancer could not meet the need of clinical application (11).
With advancing research, the recent focus of interest has shifted to the tumor microenvironment (TME) (12). The tumor microenvironment (the internal environment in which tumor cells produce and live) includes not only the tumor cells themselves but also peripheral fibroblasts, immune and inflammatory cells, and other various cells. Meanwhile, the cellular interstitium, microvessels, and biomolecules infiltrated in nearby areas are characterized by hypoxia, chronic inflammation, and immunosuppression (13,14). The involvement of immune cells in the development of cancer has also been reported by many studies (15). The expanding scale and inherent complexity of biological data have encouraged a growing use of machine learning to build informative and predictive models of the underlying biological processes (16). In this study, potential colorectal cancer diagnostic genes were screened by using machine learning methods. To construct a more accurate diagnostic signature, we employed two most commonly used traditional machine learning methods, Least Absolute Shrinkage and Selection Operator (LASSO) and Support Vector Machine (SVM) algorithms (16,17). Additionally, immune cell infiltration was investigated by CIBERSORT analysis (18) to observe the correlation between key genes and infiltrating immune cells to identify new biomarkers for colorectal cancer diagnosis and subsequent treatment. We present the following article in accordance with the STARD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-536/rc).
Methods
Study design
This is a bioinformatics analysis study and the potential colorectal cancer diagnostic genes were screened by using machine learning methods. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Download, standardization, and integration of data
Two datasets—GSE41328 and GSE106582—were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). GSE41328 and GSE106582 are gene expression datasets of 20 cases of colorectal cancer tissues and 194 cases of normal tissues, respectively. The data were normalized and merged with the R software packages “limma” and “Sva” as the training set. GSE110225 is a gene expression dataset of 60 cases of colorectal adenocarcinoma and normal tissues, which was used as the validation set for key gene differences.
Differential gene analysis
The gene expression differences between colon cancer and normal tissues were analyzed, and the screening threshold for differentially expressed genes (DEGs) was |logFC|=2 adj.P.Val. Filter <0.05. Volcano maps and heat maps were drawn with the “ggplot2” and “pheatmap” software packages. Ggplot2 (19) and pheatmap are R software packages in R language that can visualize gene expression in normal and tumor groups (20).
Gene Ontology (GO) annotation, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway clustering, Disease Ontology (DO), cluster analysis, and Gene Set Enrichment Analysis (GSEA) of differential genes
Gene cluster or enrichment analysis was employed to cluster various functions, pathways, and disease-associated genes, using the R software packages “org.Hs.eg.db” and “enrichplot”. Each GO annotation consists of a gene and the corresponding GO term, which mainly includes three aspects: molecular functions (MF), biological process (BP), and cellular components (CC) (21). The KEGG database is a bioinformatics database established in 1995 by the Kanehisa laboratory at the Bioinformatics Center, Kyoto University, Japan. It is now an important bioinformatics knowledge base for integrating and interpreting large-scale molecular datasets generated by genome sequencing and other high-throughput experimental techniques. The most central database is the KEGG PATHWAY and KEGG ORTHOLOGY database (22). The KEGG clustering pathway of differential genes was applied in this study. DO analysis is a simple analysis of genetic disease enrichment (23), which was performed by using the “DOSE” R software package. The condition for the GO entries annotation, KEGG pathway, and disease analysis in this study was adj.P.Val. Filter <0.05. GSEA enrichment analysis was performed on the results of the KEGG and GO analyses by the data sets c2.cp.kegg.v7.4.symbols.gmt and c5.go.v7.4.symbols.gmt, respectively.
Screening and validation of the key colon cancer genes by LASSO and SVM regression
LASSO and SVM regression are two machine learning methods commonly used to screen variables (24,25); two regressions of the selected differential genes were intersected as key diagnostic colorectal cancer genes in this study. The diagnostic ability of the key colorectal cancer genes was examined using the area under the receiver operating characteristic (ROC) curve (AUC). Using gene set GSE110225 as the training set, the differences in the expression of the key tumor genes and their diagnostic ability for colon cancer were observed. A value of 0.75 was deemed as useful discrimination performance of AUC (26).
Analysis of immune cell infiltration in patients with colon cancer
CIBERSORT is a tool for deconvolving the expression matrix of immune cell subtypes based on the principle of linear support vector regression of genes. Immune cell infiltration was estimated by RNA-Seq data first published in Nature Methods in 2015 (27), which is currently the most commonly used analytical tool for immune cell infiltration estimation (18). The relative quantity of infiltrating immune cells in patients with colon cancer and the correlation between immune cells and the key diagnostic colorectal cancer genes were analyzed by CIBERSORT in this study. The correlation between the 11 key genes and immune cells was represented by a lollipop chart.
Statistical analysis
The gene expression differences between colon cancer and normal tissues were screened by LASSO and SVM regression based on machine learning method. The diagnostic performance of the genes was assessed using AUC. The distribution of the differentially expressed genes was shown by heatmaps. A two-tailed P value <0.05 was considered as statistical significance. All the statistical analyses were performed by using R software (Version 4.1.1).
Results
Analysis results of expressed genes in colon cancer tissues and normal tissues
The datasets were downloaded, merged, and normalized, followed by gene difference analysis, as shown in Figure 1. Filtered with the condition of |logFC|>2, the DEG analysis revealed 60 differentially expressed genes between colon cancer tissues and normal tissues, including 43 downregulated and 17 upregulated genes (Figure 2A). In colorectal cancer patients, the upregulated genes included CLDN1, FOXQ1, and TRIB3, and the downregulated genes included CA1, CLCA4, and AQP8. The specific results are shown in Figure 2B.
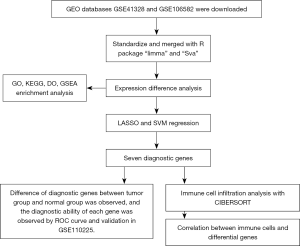
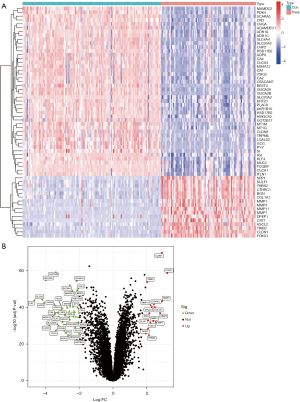
GO annotation, clustering KEGG pathway, and GSEA analysis of the differential genes
GO annotation and KEGG pathway clustering analyses were performed on the differential genes (Figure 3A,3B). The GO annotation demonstrated that the BPs of the key genes were clustered in ‘extracellular matrix organization’, ‘extracellular encapsulating structure organization’, and ‘collagen metabolic process’; the cellular components clustered in the ‘apical part of cell’, ‘cell membrane projection’, and ‘cluster of actin-based cell projections’; the molecular functions clustered in ‘metallopeptidase activity’, ‘oxidoreductase activity’, and ‘cyclase regulator activity’; KEGG showed that the key genes clustered in ‘bile secretion’, ‘nitrogen metabolism’, and ‘retinol metabolism’ on the pathway. The GO annotation and KEGG results were further analyzed by GSEA, and in colorectal cancer patients the pathway clustered in KEGG-CELL-CYCLE, KEGG-DNA-REPLICATION, and KEGGPROTEEASOME. GO entries were clustered in GOBP_CHROMOSOME_SEGREGATION, GOBP_DNA_CONFORMATION_CHANGE, and GOBP_DNA_REPAIR. The specific results are shown in Figure S1.
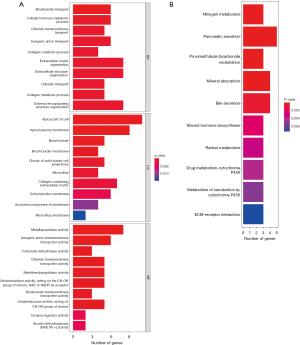
Screening and validation of the key colon cancer genes by LASSO and SVM regression
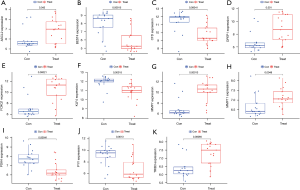
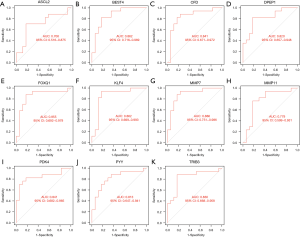
LASSO and SVM regression identified 14 and 19 genes associated with a diagnosis of colorectal cancer, respectively, for which the intersection was taken (see Table 1). Eventually, 11 genes were screened as the key diagnostic genes for colorectal cancer (Figure 4A-4C), including ASCL2, BEST4, CFD, DPEPCFD, FOXQ1, KLF4, MMP7, MMP11, PYY, PDK4, and TRIB3. The AUCs of the 11 key genes associated with colon cancer were 91.4%, 96.0%, 93.2%, 91.6%, 97.3%, 96.6%, 97.2%, 95.5%, 93.6%, 95.3%, and 97.4%, respectively (Figure 4D). The 11 key genes were significantly different in expression in colorectal cancer and normal tissues in the validation set GSE110225 (P<0.05), with ASCL2, DPEPCFD, FOXQ1, MMP7, MMP11, and TRIB3 being highly expressed in colon cancer patients (Figure 5). The AUCs of the 11 key diagnostic genes for colorectal cancer in the validation set were 70.6%, 86.2%, 84.1%, 82.0%, 85.5%, 86.2%, 88.6%, 77.9%, 84.1%, 81.3%, and 83.0%, respectively (Figure 6).
Table 1
| Different genes of LASSO regression | Different genes of SVM regression | Intersection genes |
|---|---|---|
| FOXQ1 | CLDN1 | FOXQ1 |
| TRIB3 | TRIB3 | TRIB3 |
| KLF4 | FOXQ1 | KLF4 |
| BEST4 | SCARA5 | BEST4 |
| MMP7 | MAMDC2 | MMP7 |
| MMP11 | ASCL2 | MMP11 |
| ASCL2 | BEST4 | ASCL2 |
| PYY | CST1 | PYY |
| CFD | GUCA2B | CFD |
| MT1M | KLF4 | PDK4 |
| PDK4 | MMP7 | DPEP1 |
| LGALS2 | MMP11 | |
| SPP1 | GUCA2A | |
| ADH1C | DPEP1 | |
| DPEP1 | CFD | |
| IGJ | CTHRC1 | |
| PYY | ||
| PDK4 | ||
| CA2 |
LASSO, Least Absolute Shrinkage and Selection Operator; SVM, Support Vector Machine.
Analysis of immune-infiltrating cells
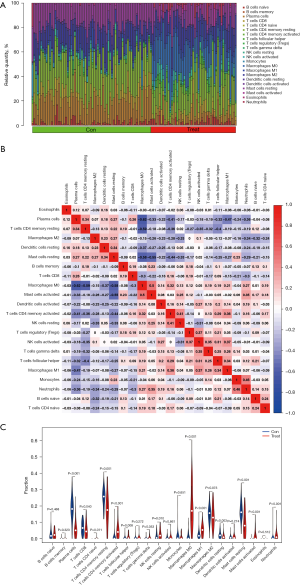
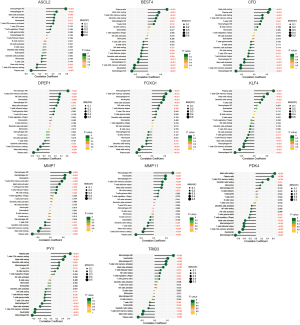
The relative quantity of immune cells in colon cancer tissues and normal tissues (Figure 7A), the correlation between infiltrating immune cells (Figure 7B), and the difference in the quantity of infiltrating immune cells between the two groups (Figure 7C) were analyzed by CIBERSORT. For the infiltrating immune cells, macrophages MO were negatively correlated with plasma cells, T cells CD4 memory were negatively correlated with Mast cells resting, which were positively correlated with Mast cells activation, with correlation coefficients of −0.62, −0.59, −0.58, and 0.50, respectively; Mast cells resting were negatively correlated with Mast cells activated, with a correlation coefficient of −0.55; The cells with different quantity of infiltrating immune cells between the two groups were plasma cells, T cells, B cells naive, NK cells resting, macrophages MO, M1, Dendritic cells resting, Mast cells resting, Mast cells activation, and Neutrophils. Genes upregulated in colorectal cancer tissues ASCL2, DPEPCFD, FOXQ1, MMP7, MMP11, and TRIB3 were associated with B cell naive, natural killer (NK) cells resting, macrophages MO, and M1 immune cells (Figure 8).
Discussion
DEG screening and GO, KEGG, and GSEA enrichment analyses were performed in this study. In total, 60 DEGs were selected, including 17 upregulated and 43 downregulated genes. The results of the GO analysis showed that DEGs were involved in ‘extracellular matrix organization’, ‘extracellular encapsulation structural organization’, ‘collagen metabolic process’, ‘cell apex’, ‘cell projection membrane population’, ‘actin-based cell projection’, ‘metallopeptidase activity’, ‘oxidoreductase activity’ and ‘cell cycle regulator’. The KEGG pathway enrichment analysis showed correlations in bile secretion, nitrogen metabolism, retinol metabolism, and extracellular matrix (ECM)-receptor interaction pathways. These pathways may play an important role in tumor immune escape, adhesion, degradation, motility, and proliferation processes (28), and their role in other cancers has been demonstrated: ECM has been shown to be upregulated in prostate cancer tissues (29), and the ECM-receptor interaction pathway was involved in the invasion and metastasis of gastric cancer (30). Additionally, a recent study on glioblastoma, the most lethal adult brain tumor, showed that the pathological features of abnormal neovascular development, diffuse tumor cell infiltration, and interactions between ECM and the glioblastoma microenvironment were important factors in disease progression (31). It was notable that the immune-related functions were found in the KEGG pathway. These results suggested that DEGs are highly relevant to the immune system, confirming our hypothesis. They may prevent immune cells from attacking cancer cells and promote immune escape to induce tumor progression and metastasis. Our GSEA enrichment analysis showed the involvement of cell cycle and DNA replication processes, suggesting that cell cycle checkpoint inhibitors or cycle arrest may be effective in treating colorectal cancer.
This is the first study to combine LASSO and Support Vector Machine-Recursive Feature Elimination (SVM-RFE) algorithms to identify and validate key biomarkers of colorectal cancer in a test set. Finally, 11 key genes were identified, including ASCL2, BEST4, CFD, DPEPCFD1, FOXQ1, KLF4, MMP7, MMP11, PYY, PDK4, and TRIB3. The AUC value was >0.91 for all 11 key genes in the training set. However, the AUC only reached about 0.7 in the validation set, which indicates that while the constructed model has a robust validation performance, its test performance needs further improvement.
Achaete scute-like-2 (ASCL2) is a key downstream molecule of the Wnt/β-catenin signaling pathway; it is a basic helix-loop helical transcription factor homolog found in enterocytes that may play an indispensable role in the maintenance of intestinal stem cell effects (32). It has been shown that ASCL2 expression leads to tumor growth arrest through miRNA-302b-mediated conditional reprogramming cells (CRC) progenitor cells and induces miR-200 expression, which further promotes the plasticity of epithelial-mesenchymal transition-mesenchymal-epithelial transition (EMT-MET) through transcriptional mechanisms (33). Downregulating ASCL2 can also promote apoptosis by enhancing autophagy in colorectal cancer cells (34).
The Bestrophin (BEST) family are newly discovered genes encoding ion channels that can function as Cl channels, HCO3 channels, or voltage-gated Ca2+ channels. BEST4 is mainly expressed within the human colon (35). BEST4 expression has been shown to be upregulated in clinical colorectal cancer samples, and its high expression level has been associated with advanced TNM stage, lymph node metastasis, and poor survival, with a potential oncogenic role in colorectal carcinogenesis and metastasis through modulation of BEST4/PI3K/Akt signal transduction (36).
Currently, there are few reports about CFD (Complement Factor D). Lipoprotein (Complement Factor D) is an adiponectin, which is mainly secreted by adipocytes. This effect is mediated by C3a, a downstream product of adiponectin, which is produced in the replacement pathway of the Complement system (37). In the literature, CFD acts as an enhancing dose for tumor proliferation and cancer stem cell (CSC) properties in breast cancer. The role of CFD in colorectal cancer remains to be further explored (38).
Dipeptidase (DPEPCFD) 1 is a zinc-dependent metalloprotease underlying glutathione and leukotriene metabolism. In colorectal cancer samples, DPEPCFD 1 expression was significantly increased in tumor tissue samples, and an elevated DPEPCFD1 mRNA expression was associated with positive lymph node metastasis (39). Additionally, DPEPCFD1 was demonstrated to promote the proliferation of colon cancer cells in vitro and in vivo through a DPEPCFD1/MYC positive feedback loop (40). The enrichment in the ECM-receptor interaction pathway found by GSEA in this study was similar to that reported in the literature. DPEPCFD1 has also been shown to play a role in rectal cancer metastasis by inhibiting the leukotriene D4 signaling pathway and increasing E-cadherin expression (41).
FOXQ1 belongs to the FOX transcription factor superfamily, characterized by a conserved binding of 110 amino acids responsible for DNA binding involved in tumor proliferation, apoptosis, migration, and invasion (42). Knockdown of FOXQ induces inhibition of cell proliferation, as well as migration, and invasion of colorectal cancer cells (43).
KLF4 family members are expressed in many cell lineages and play crucial roles in development, metabolism, and multipotency. Their dysregulation is highly involved in the development of human diseases, including cancer, and they play an important role in regulating intestinal epithelial homeostasis (44). KLF4 promotes tumor development by epigenetic modification, and the increased expression of miR-29a has been shown to promote colorectal cancer metastasis by directly targeting KLF4 to regulate MMP2/E-cad (45). Furthermore, KLF4 protein expression has been shown to correlate significantly with colorectal cancer differentiation in clinical specimens by immunohistochemistry, and downregulation of KLF4 expression may contribute to poor tumor differentiation (46).
Matrix metalloproteinase (MMP) is an enzyme component that degrades extracellular matrix proteins and promotes cancer invasion and metastasis. MMPs have been studied in serum and tissues, and an increased expression of specific MMPs has been associated with poor prognostic parameters (47). Wu et al. found that MMP7 expression was associated with colorectal cancer metastasis and poor prognosis (48). In addition, it was found that MPC1 mediated MMP7 activation of the Wnt/β-catenin pathway by promoting β-catenin nuclear translocation after silencing (49). MMP11 has also been shown to be associated with poor prognosis in gastric cancer (50).
Peptide YY, originally isolated from the pig intestine, is restricted to endocrine cells in the colon. A comprehensive bioinformatics analysis exploring the clinical value of primary CRC biomarkers found that PYY was a core gene (51). However, it has been concluded that PYY is unlikely to be involved in the development and growth of colorectal cancer. Whether this conclusion remains valid should be confirmed by subsequent experimental validation (52).
Pyruvate Dehydrogenase Kinase (PDK) generates four kinase families in humans. PDK4 is mainly expressed in muscle and affects glucose consumption during metabolism (53). It has been reported in the literature that when PDK4 was stably inhibited, colorectal cancer cell migration and invasion were reduced, and apoptosis was increased. PDK4 also reduced the expression of vimentin, hypoxia-inducible factor-1 (HIF-1), and vascular endothelial growth factor A (VEGFA) (54).
Tribbles pseudokinase 3 (TRIB3) contains a substrate-binding domain. However, it lacks the conserved catalytic amino acid motif required for kinase activity (55). Recent studies have shown that TRIB3 is a crucial oncoprotein associated with many different types of cancer, including hepatocellular carcinoma, colorectal cancer, and gastric cancer (56-58).
To quantify the relative proportion of infiltrating immune cells in colorectal cancer gene expression profiles, immune cell infiltration can be calculated using the bioinformatics algorithm CIBERSORT, which is increasingly used to estimate immune cell infiltration because of its good performance (27). In this study, CIBERSORT was used to investigate the role of immune cell infiltration in colorectal cancer. Our analysis found differences in immune cells between the colorectal cancer and control groups. The differences were found among plasma cells, T cells CD8, T cells CD4 memory, T cells CD4 memory resting, T cells CD4 memory activated, T cells follicular helper, NK cells resting, Macrophages M0, Macrophages M1, Dendritic cells resting, Dendritic cells activated, Mast cells resting, Mast cells activated, and Neutrophils. Additionally, the expression of T cells CD4 activated, T cells follicular helper, NK cells resting, Macrophages M0, Macrophages M1, Mast cells activated, and Neutrophils were higher in the colorectal cancer group. We also studied the relationship between the expression of key genes in colorectal cancer and immune cells to provide new clinical guidance for cancer diagnosis.
Using comprehensive bioinformatics and machine learning algorithms, the genomic landscape of colorectal cancer and its correlation with immune cell infiltration was elucidated in this study. A total of 11 prognosis-associated key genes were found to play pleiotropic roles in the TME of colorectal cancer. These central genes are involved in the formation of the immune microenvironment and could represent potential therapeutic targets. Further experiments on the current findings based on retrospective datasets and clinical specimens should be performed to validate our results.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Liaoning Province (No. 2021-MS-330).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-536/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-536/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shaukat A, Kahi CJ, Burke CA, et al. ACG Clinical Guidelines: Colorectal Cancer Screening 2021. Am J Gastroenterol 2021;116:458-79. [Crossref] [PubMed]
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Wu C, Li M, Meng H, et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci 2019;62:640-7. [Crossref] [PubMed]
- Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 2000;343:78-85. [Crossref] [PubMed]
- Monahan KJ, Bradshaw N, Dolwani S, et al. Guidelines for the management of hereditary colorectal cancer from the British Society of Gastroenterology (BSG)/Association of Coloproctology of Great Britain and Ireland (ACPGBI)/United Kingdom Cancer Genetics Group (UKCGG). Gut 2020;69:411-44. [Crossref] [PubMed]
- Li D, Zhang L, Fu J, et al. SCTR hypermethylation is a diagnostic biomarker in colorectal cancer. Cancer Sci 2020;111:4558-66. [Crossref] [PubMed]
- Wu Y, Yang X, Jiang G, et al. 5'-tRF-GlyGCC: a tRNA-derived small RNA as a novel biomarker for colorectal cancer diagnosis. Genome Med 2021;13:20. [Crossref] [PubMed]
- Li N, Li J, Mi Q, et al. Long non-coding RNA ADAMTS9-AS1 suppresses colorectal cancer by inhibiting the Wnt/β-catenin signalling pathway and is a potential diagnostic biomarker. J Cell Mol Med 2020;24:11318-29. [Crossref] [PubMed]
- Maurya NS, Kushwaha S, Chawade A, et al. Transcriptome profiling by combined machine learning and statistical R analysis identifies TMEM236 as a potential novel diagnostic biomarker for colorectal cancer. Sci Rep 2021;11:14304. [Crossref] [PubMed]
- Dariya B, Aliya S, Merchant N, et al. Colorectal Cancer Biology, Diagnosis, and Therapeutic Approaches. Crit Rev Oncog 2020;25:71-94. [Crossref] [PubMed]
- Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960-4. [Crossref] [PubMed]
- Galon J, Fridman WH, Pagès F. The adaptive immunologic microenvironment in colorectal cancer: a novel perspective. Cancer Res 2007;67:1883-6. [Crossref] [PubMed]
- Hu H, Krasinskas A, Willis J. Perspectives on current tumor-node-metastasis (TNM) staging of cancers of the colon and rectum. Semin Oncol 2011;38:500-10. [Crossref] [PubMed]
- Ogino S, Giannakis M. Immunoscore for (colorectal) cancer precision medicine. Lancet 2018;391:2084-6. [Crossref] [PubMed]
- Greener JG, Kandathil SM, Moffat L, et al. A guide to machine learning for biologists. Nat Rev Mol Cell Biol 2022;23:40-55. [Crossref] [PubMed]
- Su Y, Tian X, Gao R, et al. Colon cancer diagnosis and staging classification based on machine learning and bioinformatics analysis. Comput Biol Med 2022;145:105409. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Ito K, Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol 2013;2:e79. [Crossref] [PubMed]
- Yao S, Liu T. Analysis of differential gene expression caused by cervical intraepithelial neoplasia based on GEO database. Oncol Lett 2018;15:8319-24. [Crossref] [PubMed]
- Wimalanathan K, Friedberg I, Andorf CM, et al. Maize GO Annotation-Methods, Evaluation, and Review (maize-GAMER). Plant Direct 2018;2:e00052. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Schriml LM, Mitraka E, Munro J, et al. Human Disease Ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res 2019;47:D955-62. [Crossref] [PubMed]
- Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997;16:385-95. [Crossref] [PubMed]
- Huang S, Cai N, Pacheco PP, et al. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genomics Proteomics 2018;15:41-51. [PubMed]
- Alba AC, Agoritsas T, Walsh M, et al. Discrimination and Calibration of Clinical Prediction Models: Users' Guides to the Medical Literature. JAMA 2017;318:1377-84. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Majoor BC, Boyce AM, Bovée JV, et al. Increased Risk of Breast Cancer at a Young Age in Women with Fibrous Dysplasia. J Bone Miner Res 2018;33:84-90. [Crossref] [PubMed]
- Andersen MK, Rise K, Giskeødegård GF, et al. Integrative metabolic and transcriptomic profiling of prostate cancer tissue containing reactive stroma. Sci Rep 2018;8:14269. [Crossref] [PubMed]
- Yan P, He Y, Xie K, et al. In silico analyses for potential key genes associated with gastric cancer. PeerJ 2018;6:e6092. [Crossref] [PubMed]
- Cui X, Morales RT, Qian W, et al. Hacking macrophage-associated immunosuppression for regulating glioblastoma angiogenesis. Biomaterials 2018;161:164-78. [Crossref] [PubMed]
- Yang Q, Huang G, Li L, et al. Potential Mechanism of Immune Evasion Associated with the Master Regulator ASCL2 in Microsatellite Stability in Colorectal Cancer. J Immunol Res 2021;2021:5964752. [Crossref] [PubMed]
- Tian Y, Pan Q, Shang Y, et al. MicroRNA-200 (miR-200) cluster regulation by achaete scute-like 2 (Ascl2): impact on the epithelial-mesenchymal transition in colon cancer cells. J Biol Chem 2014;289:36101-15. [Crossref] [PubMed]
- Wang H, Ye T, Cai Y, et al. Downregulation of Ascl2 promotes cell apoptosis by enhancing autophagy in colorectal cancer cells. J Gastrointest Oncol 2021;12:630-8. [Crossref] [PubMed]
- Ito G, Okamoto R, Murano T, et al. Lineage-specific expression of bestrophin-2 and bestrophin-4 in human intestinal epithelial cells. PLoS One 2013;8:e79693. [Crossref] [PubMed]
- He XS, Ye WL, Zhang YJ, et al. Oncogenic potential of BEST4 in colorectal cancer via activation of PI3K/Akt signaling. Oncogene 2022;41:1166-77. [Crossref] [PubMed]
- Goto H, Shimono Y, Funakoshi Y, et al. Adipose-derived stem cells enhance human breast cancer growth and cancer stem cell-like properties through adipsin. Oncogene 2019;38:767-79. [Crossref] [PubMed]
- Mizuno M, Khaledian B, Maeda M, et al. Adipsin-Dependent Secretion of Hepatocyte Growth Factor Regulates the Adipocyte-Cancer Stem Cell Interaction. Cancers (Basel) 2021;13:4238. [Crossref] [PubMed]
- Tachibana K, Saito M, Imai JI, et al. Clinicopathological examination of dipeptidase 1 expression in colorectal cancer. Biomed Rep 2017;6:423-8. [PubMed]
- Liu Q, Deng J, Yang C, et al. DPEP1 promotes the proliferation of colon cancer cells via the DPEP1/MYC feedback loop regulation. Biochem Biophys Res Commun 2020;532:520-7. [Crossref] [PubMed]
- Park SY, Lee SJ, Cho HJ, et al. Dehydropeptidase 1 promotes metastasis through regulation of E-cadherin expression in colon cancer. Oncotarget 2016;7:9501-12. [Crossref] [PubMed]
- Yang M, Liu Q, Dai M, et al. FOXQ1-mediated SIRT1 upregulation enhances stemness and radio-resistance of colorectal cancer cells and restores intestinal microbiota function by promoting β-catenin nuclear translocation. J Exp Clin Cancer Res 2022;41:70. [Crossref] [PubMed]
- Weng W, Okugawa Y, Toden S, et al. FOXM1 and FOXQ1 Are Promising Prognostic Biomarkers and Novel Targets of Tumor-Suppressive miR-342 in Human Colorectal Cancer. Clin Cancer Res 2016;22:4947-57. [Crossref] [PubMed]
- Taracha-Wisniewska A, Kotarba G, Dworkin S, et al. Recent Discoveries on the Involvement of Krüppel-Like Factor 4 in the Most Common Cancer Types. Int J Mol Sci 2020;21:8843. [Crossref] [PubMed]
- Tang W, Zhu Y, Gao J, et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br J Cancer 2014;110:450-8. [Crossref] [PubMed]
- Hu R, Zuo Y, Zuo L, et al. KLF4 Expression Correlates with the Degree of Differentiation in Colorectal Cancer. Gut Liver 2011;5:154-9. [Crossref] [PubMed]
- Yen JH, Chio WT, Chuang CJ, et al. Improved Wound Healing by Naringin Associated with MMP and the VEGF Pathway. Molecules 2022;27:1695. [Crossref] [PubMed]
- Wu Q, Yang Y, Wu S, et al. Evaluation of the correlation of KAI1/CD82, CD44, MMP7 and β-catenin in the prediction of prognosis and metastasis in colorectal carcinoma. Diagn Pathol 2015;10:176. [Crossref] [PubMed]
- Tian GA, Xu CJ, Zhou KX, et al. MPC1 Deficiency Promotes CRC Liver Metastasis via Facilitating Nuclear Translocation of β-Catenin. J Immunol Res 2020;2020:8340329. [Crossref] [PubMed]
- Tian X, Ye C, Yang Y, et al. Expression of CD147 and matrix metalloproteinase-11 in colorectal cancer and their relationship to clinicopathological features. J Transl Med 2015;13:337. [Crossref] [PubMed]
- Wang YR, Meng LB, Su F, et al. Insights regarding novel biomarkers and the pathogenesis of primary colorectal carcinoma based on bioinformatic analysis. Comput Biol Chem 2020;85:107229. [Crossref] [PubMed]
- El-Salhy M, Mazzawi T, Gundersen D, et al. The role of peptide YY in gastrointestinal diseases and disorders Int J Mol Med 2013;31:275-82. (review). [Crossref] [PubMed]
- Kim CJ, Terado T, Tambe Y, et al. Cryptotanshinone, a novel PDK 4 inhibitor, suppresses bladder cancer cell invasiveness via the mTOR/β catenin/N cadherin axis. Int J Oncol 2021;59:40. [Crossref] [PubMed]
- Leclerc D, Pham DN, Lévesque N, et al. Oncogenic role of PDK4 in human colon cancer cells. Br J Cancer 2017;116:930-6. [Crossref] [PubMed]
- Clark RA. The trouble with TRIBbles: TRIB3 blocks CD8 T cell homing to colorectal cancers. Sci Immunol 2022;7:eabo2990. [Crossref] [PubMed]
- Liu C, Zhang W, Wang J, et al. Tumor-associated macrophage-derived transforming growth factor-β promotes colorectal cancer progression through HIF1-TRIB3 signaling. Cancer Sci 2021;112:4198-207. [Crossref] [PubMed]
- Liu S, Ni C, Li Y, et al. The Involvement of TRIB3 and FABP1 and Their Potential Functions in the Dynamic Process of Gastric Cancer. Front Mol Biosci 2021;8:790433. [Crossref] [PubMed]
- Örd T, Örd D, Kaikkonen MU, et al. Pharmacological or TRIB3-Mediated Suppression of ATF4 Transcriptional Activity Promotes Hepatoma Cell Resistance to Proteasome Inhibitor Bortezomib. Cancers (Basel) 2021;13:2341. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


