
A case report of malignant neuroectodermal tumor of the gastrointestinal tract without common gene fusion in a soft tissue tumor
Introduction
Malignant gastrointestinal neuroectodermal tumor (GNET), which is also known as clear cell sarcoma (CCS)-like tumor of the gastrointestinal tract (CCSLTGT), is a rare type of malignant soft tissue sarcoma that mainly occurs in the wall of the gastrointestinal tract (1). To date, <50 articles on CCSLTGTs have been published. This type of tumor mainly affects young people, and it has a poor prognosis and a controversial origin. Because of its similarity to CCS in terms of its histology, immunohistochemistry, and gene fusion (2), GNET is histologically and immunohistochemically similar to many sarcomas, especially clear cell sarcoma (CCS). (3). And more and more evidence supports that GNET and CCS were different kind of tumors (4). To the best of our knowledge, CCSLTGT GNET has only seldomly been diagnosed in the absence of EWSR1-CREB1 or EWSR1-ATF1 gene fusions (3,4). Here, we presented a case of hepatic metastases from GNETs negative for common gene fusion in a soft tissue tumor. This case will provide more evidence for the diagnosis of EWSR1 fusion-negative GNET in clinic. We present the following case in accordance with the CARE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-387/rc).
Case presentation
Ethical statement
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Clinical history
A 48-year-old woman experienced melena accompanied by dizziness and fatigue for >15 days. A gastroscopic examination at another hospital revealed a mass in the gastric antrum with bleeding at the center. The patient initially refused surgery and received only symptomatic treatment, including the maintenance of hemostasis and the administration of gastrointestinal medications and infusions. A few days after discharge, the patient’s symptoms of melena, dizziness, and fatigue worsened. On admission to our hospital, her blood pressure was 92/56 mmHg. Routine blood tests revealed a low hemoglobin level (6.4 gm/dL); however, her tumor index was normal.
Gastroscopy was performed again after admission (see Figure 1A). An abdominal computed tomography (CT) scan showed a mass arising from the stomach with hepatic metastasis. Chest CT showed local rib metastasis. After admission, the patient's hemoglobin level reduced from 6.4 to 5.5 gm/dL, and symptoms of anemia were observed. We administered 200 mL of a red-blood cell suspension to the patient 4 times, and her anemia symptoms improved. Radical gastrectomy with resection of the liver metastasis was performed 5 days later. The gross operative findings included the stomach and segment VI of the liver. The gastric tumor was located in the posterior wall of the gastric antrum, approximately 1.5 cm from the pylorus. The excised tumor had a round shape and measured approximately 5 cm in diameter. There was a 5-cm diameter mass in segment VI of the liver. No mass or fluid accumulation was observed in other organs in the abdominal cavity. There were no family members with a disease similar to the patient’s disease.
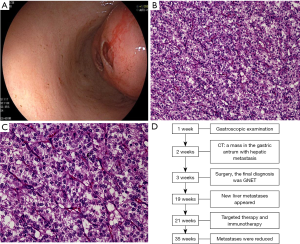
Pathology
The surgical specimens were fixed with 10% neutral buffered formalin, 40 g/L of formaldehyde buffer, and embedded in paraffin, as per the routine process. At the microscopic level, the tumor, in which a local ulcer had formed, was observed in the submucosa and muscularis propria of the stomach wall. The tumor had medium epithelioid cells, and exhibited a nested growth pattern. Most of the tumor cells had round or oval nuclei with small nucleoli surrounded by moderate amounts of eosinophilic cytoplasm. Additionally, multinucleated osteoclast-like giant cells were found in the tumor (see Figure 1B,1C). However, there was no tumor invasion at the surgical margin and hepatic serosa, and there were no tumor cells in the lymph nodes. Further, the tumor cells in the liver were similar to those in the stomach. The timeline of the patient’s diagnosis, treatment and progression was showed in Figure 1D.
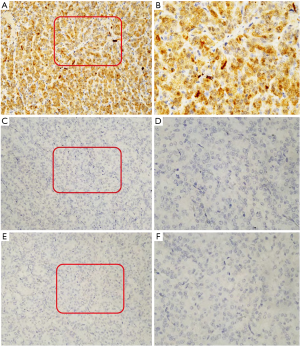
Immunohistochemical studies were performed on representative paraffin specimens. The following commercial immunohistochemical antibodies were listed: CK-pan, CgA, SYN, desmin, MyOD1, TTF-1, napsin-A, DOG1, CDX2, AFP, hepatocyte, Arginase-1, Cam5.2, HMB45, MelanA, vimentin, SMA, and SOX10 were obtained from Guangzhou Lbp Medicine Science & Technology Company Limited. CD99 and CD117 were obtained from Fuzhou Maixin Biotechnology Development Company Limited. CK7, CK20, and Ki-67 were obtained from Roche Life Science Company. Among them, SYN, CD34, vimentin, S-100, SOX10, CD56, TTF-1, CgA, CDX2 and desmin were analyzed on the Dako-link 48 platform. CD99, Napsin, SMA, Melan-A, HMB-45, AFP, Arginase-1, and hepatocyte were analyzed on the Dako-omnis platform. Ki-67 and Cam5.2 were analyzed on the Roche-XT platform. Immunohistochemical staining showed that the tumor cells were positive for S100 protein, Ki-67 (the positive rate of Ki-67 is 40%), CD56, and vimentin, focally positive for CD34 and SMA, and negative for CD117, CK-pan, CK7, CK20, chromogranin A, synaptophysin, desmin, MyOD1, TTF-1, napsin-A, DOG1, CDX2, AFP, hepatocyte, Arginase-1, Cam5.2, CD99, HMB-45, Melan-A, and SOX-10 (see Figure 2A-2F). Based on the histological and immunohistochemical findings, GNET was diagnosed.
EWSR1 gene fusion is common in GNET. To confirm the GNET diagnosis in our case, we analyzed 200 interphase tumor cells in at least 2 fields by fluorescence in situ hybridization using the EWSR1 gene probe (obtained from Guangzhou Lbp Medicine Science and Technology Company Limited). No rearrangements in EWSR1 were detected (see Figure 3A). Next, we detected 64 pairs of common soft tissue tumor fusion genes using next-generation sequencing, including multiplex polymerase chain reaction. The detected gene panels are listed in Table 1. We extracted total ribonucleic acid (RNA) from paraffin tissue using the magnetic bead method, amplified soft tissue tumor-related fusion genes using an Ampliseq RNA SARC fusion panel, and performed amplicon library construction and sequencing experiments (using the Ion PGM™ system kit). We further tested for other mutations, including single nucleotide polymorphisms (SNPs), insertions and deletions, base deletions of <50 bp, somatic mutations, predisposing genes, and driver mutations, in the tumor genes using whole-exome sequencing more comprehensively. We compared these results with those obtained from a series of databases (see Figure 3B).
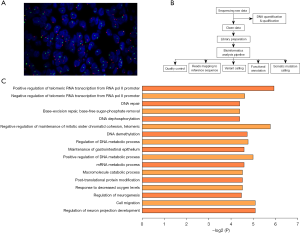
Table 1
| Fusion gene | 5' gene transcript number | 5' genetic testing range | 3' gene transcript number | 3' genetic testing range |
|---|---|---|---|---|
| ACTB-GLI1 | NM_001101.3 | Exon2/3 | NM_005269.3 | Exon5-7 |
| ASPSCR1-TFE3 | NM_024083.4 | Exon7 | NM_006521.6 | Exon5/6 |
| ATIC-ALK | NM_004044.7 | Exon7 | NM_004304.5 | Exon20 |
| BCOR-CCNB3 | NM_017745.6 | Exon15 | NM_033031.2 | Exon5 |
| CARS-ALK | NM_001751.6 | Exon17 | NM_004304.5 | Exon20 |
| CDH11-USP6 | NM_001797.4 | Exon1/2 | NM_004505.4 | Exon1/2 |
| CIC-DUX4 | NM_005258673.2 | Exon20 | NM_033178.4 | Exon1 |
| CIC-FOXO4 | NM_005258673.2 | Exon19/20 | NM_005938.4 | Exon2 |
| CLTC-ALK | NM_004859.4 | Exon30 | NM_004304.5 | Exon20 |
| COL1A1-PDGFB | NM_000088.4 | Full coding region | NM_002608.4 | Exon2 |
| COL1A1-USP6 | NM_000088.4 | Exon1 | NM_004505.4 | Exon1/2 |
| COL1A2-PLAG1 | NM_000089.4 | Exon1 | NM_002655.3 | Exon2/3 |
| CNBP-USP6 | NM_003418.5 | Exon1 | NM_004505.4 | Exon1/2 |
| EPC1-PHF1 | NM_025209.5 | Exon10 | NM_024165.3 | Exon2 |
| ETV6-NTRK3 | NM_001987.5 | Exon4/5 | NM_002530.4 | Exon15 |
| EWSR1-ATF1 | NM_005243.4 | Exon5-10 | NM_005171.5 | Exon4/5/7-10 |
| EWSR1-CREB1 | NM_005243.4 | Exon7 | NM_134442.5 | Exon7 |
| EWSR1-DDIT3 | NM_005243.4 | Exon7/9/10/13 | NM_001195057.1 | Exon2 |
| EWSR1-ERG | NM_005243.4 | Exon6/7/9/10 | NM_004449.4 | Exon8-11 |
| EWSR1-ETV1 | NM_005243.4 | Exon7 | NM_004956.5 | Exon12 |
| EWSR1-ETV4 | NM_005243.4 | Exon7 | NM_001079675.5 | Exon8/9 |
| EWSR1-FEV | NM_005243.4 | Exon7/9/10 | NM_017521.3 | Exon2 |
| EWSR1-FLI1 | NM_005243.4 | Exon7-10 | NM_002017.5 | Exon4-9 |
| EWSR1-NFATC2 | NM_005243.4 | Exon8 | NM_173091.4 | Exon3 |
| EWSR1-NR4A3 | NM_005243.4 | Exon11-13 | NM_006981.4 | Exon1-3 |
| EWSR1-PBX1 | NM_005243.4 | Exon7/8 | NM_002585.4 | Exon5 |
| EWSR1-POU5F1 | NM_005243.4 | Exon6/7 | NM_002701.6 | Exon2 |
| EWSR1-PATZ1 | NM_005243.4 | Exon8 | NM_032051.2 | Exon1 |
| EWSR1-SMARCA5 | NM_005243.4 | Exon7 | NM_003601.4 | Exon5 |
| EWSR1-SP3 | NM_005243.4 | Exon7/8 | NM_003111.4 | Exon6 |
| EWSR1-WT1 | NM_005243.4 | Exon8-10 | NM_000378.6 | Exon8-10 |
| EWSR1-ZNF444 | NM_005243.4 | Exon8 | NM_018337.4 | Exon5 |
| FUS-ATF1 | NM_004960.4 | Exon5 | NM_005171.5 | Exon5 |
| FUS-CREB3L2 | NM_004960.4 | Exon5-7 | NM_194071.4 | Exon5/6 |
| FUS-DDIT3 | NM_004960.4 | Exon3/5-9/11-13 | NM_001195057.1 | Exon2/3 |
| FUS-ERG | NM_004960.4 | Exon5-8 | NM_004449.4 | Exon8-11 |
| HAS2-PLAG1 | NM_005328.3 | Exon1 | NM_002655.3 | Exon2 |
| HEY1-NCOA2 | NM_012258.4 | Exon4 | NM_006540.4 | Exon13 |
| HMGA2-LPP | NM_003483.6 | Exon2/3 | NM_005578.3 | Exon7/9 |
| LMNA-NTRK1 | NM_170707.4 | Exon2 | NM_001007792.1 | Exon10 |
| MEAF6-PHF1 | NM_022756.6 | Exon5 | NM_024165.3 | Exon2 |
| MYH9-USP6 | NM_002473.6 | Exon1 | NM_001304284.2 | Exon9/10 |
| NAB2-STAT6 | NM_005967.4 | Exon2-7 | NM_003153.5 | Exon2-7/16-19 |
| OMD-USP6 | NM_005014.2 | Exon1 | NM_004505.4 | Exon1 |
| PAX3-FOXO1 | NM_181459.4 | Exon7 | NM_002015.4 | Exon2 |
| PAX3-NCOA1 | NM_181459.4 | Exon6/7 | NM_005264625.1 | Exon14/15 |
| PAX3-NCOA2 | NM_181459.4 | Exon7 | NM_006540.4 | Exon12 |
| PAX7-FOXO1 | NM_002584.3 | Exon7 | NM_002015.4 | Exon2 |
| RANBP2-ALK | NM_006267.5 | Exon18 | NM_004304.5 | Exon20 |
| SERPINE1-FOSB | NM_000602.5 | Exon1 | NM_006732.3 | Exon1/2 |
| SFPQ-TFE3 | NM_005066.3 | Exon7/9 | NM_006521.6 | Exon5/6 |
| SS18-SSX1 | NM_001007559.3 | Exon9/10 | NM_005635.4 | Exon4-6 |
| SS18-SSX2 | NM_001007559.3 | Exon10 | NM_003147.5 | Exon6 |
| SS18-SSX4 | NM_001007559.3 | Exon10 | NM_005636.3 | Exon3/6/7 |
| TAF15-NR4A3 | NM_207037.1 | Exon6/7 | NM_006981.4 | Exon3 |
| TCF12-NR4A3 | NM_207036.2 | Exon5 | NM_006981.4 | Exon3 |
| TFG-NR4A3 | NM_006070.6 | Exon7 | NM_006981.4 | Exon3 |
| THRAP3-USP6 | NM_005119.4 | Exon1 | NM_004505.4 | Exon1 |
| TPM3-ALK | NM_153649.4 | Exon6/7 | NM_004304.5 | Exon20 |
| TPM3-NTRK1 | NM_153649.4 | Exon6/7 | NM_001007792.1 | Exon10 |
| TPM4-ALK | NM_003290.2 | Exon6/7 | NM_004304.5 | Exon20 |
| TPR-NTRK1 | NM_003292.3 | Exon21 | NM_001007792.1 | Exon10 |
| WWTR1-CAMTA1 | NM_015472.6 | Exon4 | NM_015215.4 | Exon8/9 |
| ZC3H7B-BCOR | NM_017590.6 | Exon10 | NM_001123385.2 | Exon7 |
Follow up
However, 6 months postoperatively, a series of follow-up CT scans revealed an increase in the liver metastasis size, with the appearance of new metastases; the metastases on the rib did not change significantly. Targeted therapy and immunotherapy (camrelizumab 200 mg and anlotinib 12 mg) were then administered to treat the patient. The liver metastases were reduced during treatment, and no new metastases appeared.
Discussion
In this article, we described a rare case of a woman presenting with a malignant gastrointestinal neuroectodermal tumor without common gene fusion in a soft tissue tumor. To the best of our knowledge, this is the first case report of a GNET mutation analysis.
GNET was first discovered in 1985 by Alpert and Beckstead (5). Zambrano et al. (6) reported 6 cases in 2003 and identified the similarities between GNET and CCS. In 2012, Stockman et al. (3) differentiated GNET from CCS using the immunohistochemical and ultrastructural evidence of 16 cases. The question of whether CCS and GNET are distinct tumors or whether GNET is a subtype of CCS has been debated for a long time. As related research became more thorough, GNET and CCS came to be considered and are now widely accepted to be two distinct entities. Most GNETs occur in the muscularis propria of the small intestine, with focal extensions into the serosa. Similar to some soft tissue sarcomas, the diagnosis of GNET remains a clinical challenge, and its diagnosis requires histological, immunohistochemical, and molecular evidence.
There are microscopic morphological and molecular similarities between GNET and CCS. The GNET and CCS tumor cells are characteristically composed of epithelioid, polygonal, or round cells with eosinophilic cytoplasm and vesicular nuclei with prominent nucleoli. However, CD68-positive, multinucleated osteoclast-like giant cells have been reported to occur in 50% of GNET cases (1,2). Such cells provide an important clue for distinguishing GNET from CCS (1,2). We did not find any osteoclast-like giant cells in the present case.
Immunohistochemically, GNET and CCS are positive for S100. However, most GNET cases are negative for melanocytic markers, such as HMB-45 and Melan-A. Additionally, many cases of GNET have shown evidence of neuroectodermal differentiation and have often shown focal reactivities for SOX10, synaptophysin, neuron-specific enolase, neurofilament, CD56, and CD57 at the immunophenotypic level (3). Gastrointestinal stromal tumors (GISTs) should also be considered for differential diagnosis. CD117 and DOG1 markers are positive in GISTs but negative in GNETs. The multinucleated osteoclast-like giant cells are rare in GIST, while c-kit fusion is common in GISTs genetically. Additionally, while granular cell tumors are also positive for S100 and CD56, their morphology under the microscope differs. The cytoplasm of granulosa cell tumors are more abundant than those of GNETs, and the cytoplasm of granular cell tumors contain augulate bodies.
Gene analysis plays an important role in diagnosing GNET. A molecular genetic study of most GNETs has found EWSR1 gene rearrangements, usually fused with ATF1 or CREB1 (4). EWSR1 has a propensity for fusing (as the 5’ partner) with a host of different genes, with its product being a member of the FET (FUS/EWSR1/TAF15) family of transcription factors (7). More than 70% of GNETs show EWSR1-ATF1 fusions, with 20–30% of cases showing EWSR1-CREB1 fusions. However, in our case, common gene rearrangements, including EWSR1, were not identified. For EWSR1 fusion-negative GNETs, there is currently less diagnostic evidence at the genetic level, so we performed further genetic analysis in this case.
Then we used next-generation sequencing technology to improve our understanding of the genetic mutations in this case. The somatic SNP mutation findings in our case revealed 374 gene locus mutations in our patient’s tumor. By comparing widely accepted cancer-associated mutated genes in the Cancer Gene Census database, we identified 15 genes involved in the mechanisms of the tumor. The Gene Ontology (GO) pathway enrichment analysis showed that 36 genes were involved in 17 pathways that regulated tumor growth (see Figure 3C). Combined with the results of The Cancer Gene Census database and the GO pathway enrichment analysis, 11 genes in this case were involved in the occurrence and development of soft tissue sarcoma, including APEX1, ATRX, TET1, POLQ, FGFR4, MEN1, MUC4, NELL1, KMT2C, PALB2, and ROCK1.
Among these, ATRX and PALB2 mutations are important in the pathogeneses of various kinds of sarcoma, such as dedifferentiated liposarcoma and leiomyosarcoma (8,9). MUC4 encodes transmembrane mucin 4 proteins, which are overexpressed in a variety of cancers (10). In our case, MUC4 was mutated at multiple sites, and may have been associated with the proliferation and metastasis of the tumor (11). FGFR4 is associated with a poor prognosis of rhabdomyosarcoma and may be a potential therapeutic target for sarcoma (12). APEX1, TET1, NELL1, KMT2C, and ROCK1 are related to the occurrence, development, and prognosis of osteosarcoma (13-15). Alterations in the POLQ gene could contribute to dendritic cell sarcoma formation but more evidence is needed (16). MEN1 mutations can be found in pulmonary carcinosarcoma (17). These genes require further study to reveal their role in the diagnosis and mechanism of GNET occurrence and development.
Driver genes play an important role in the modification and activation of cell functions. The identification of driver genes provides important mechanistic, diagnostic, and therapeutic insights (18). We identified the driving mutations in the tumor genes of our patient and compared them to the mutant genes in the latest versions of the Cancer Gene Census database (which contained 125 mutation driver genes) (18,19). We found that 6 genes had the following missense mutations: NEB, PALB2, ATRX, PWP1, GRIN3A, and ARID1B. The ATRX and PALB2 genes are closely related to sarcomas. ARID1B regulates the expression of the SWI/SWF complex, which may be potentially related to the mechanism of GNET formation (20). This provide more valuable clues for the diagnosis of GNET; however, the role of these genes in the occurrence and progression of neoplasms has not yet been clarified.
Conclusions
We reported a case of GNET lacking any common gene fusion in soft tissue tumors and analyzed the gene mutations of GNET. To date, few studies have been conducted on this rare type of GNET, but next-generation sequencing technologies can help us understand this disease. 11 genes included known cancer genes PALB2, ATRX were mutated in the case and were closely related to the development of soft tissue sarcomas. The genes identified in our case may contribute to the study of GNET genes.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-387/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-387/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that any questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chang B, Yu L, Guo WW, et al. Malignant Gastrointestinal Neuroectodermal Tumor: Clinicopathologic, Immunohistochemical, and Molecular Analysis of 19 Cases. Am J Surg Pathol 2020;44:456-66. [Crossref] [PubMed]
- Wang J, Thway K. Clear cell sarcoma-like tumor of the gastrointestinal tract: an evolving entity. Arch Pathol Lab Med 2015;139:407-12. [Crossref] [PubMed]
- Stockman DL, Miettinen M, Suster S, et al. Malignant gastrointestinal neuroectodermal tumor: clinicopathologic, immunohistochemical, ultrastructural, and molecular analysis of 16 cases with a reappraisal of clear cell sarcoma-like tumors of the gastrointestinal tract. Am J Surg Pathol 2012;36:857-68. [Crossref] [PubMed]
- Green C, Spagnolo DV, Robbins PD, et al. Clear cell sarcoma of the gastrointestinal tract and malignant gastrointestinal neuroectodermal tumour: distinct or related entities? A review. Pathology 2018;50:490-8. [Crossref] [PubMed]
- Alpers CE, Beckstead JH. Malignant neuroendocrine tumor of the jejunum with osteoclast-like giant cells. Enzyme histochemistry distinguishes tumor cells from giant cells. Am J Surg Pathol 1985;9:57-64. [Crossref] [PubMed]
- Zambrano E, Reyes-Mugica M, Franchi A, et al. An osteoclast-rich tumor of the gastrointestinal tract with features resembling clear cell sarcoma of soft parts: reports of 6 cases of a GIST simulator. Int J Surg Pathol 2003;11:75-81. [Crossref] [PubMed]
- Flucke U, van Noesel MM, Siozopoulou V, et al. EWSR1-The Most Common Rearranged Gene in Soft Tissue Lesions, Which Also Occurs in Different Bone Lesions: An Updated Review. Diagnostics (Basel) 2021;11:1093. [Crossref] [PubMed]
- Valenzuela M, Amato R, Sgura A, et al. The Multiple Facets of ATRX Protein. Cancers (Basel) 2021;13:2211. [Crossref] [PubMed]
- Beer A, Beck R, Schedel A, et al. A rare PALB2 germline variant causing G2/M cell cycle arrest is associated with isolated myelosarcoma in infancy. Mol Genet Genomic Med 2021;9:e1746. [Crossref] [PubMed]
- Bhatia R, Gautam SK, Cannon A, et al. Cancer-associated mucins: role in immune modulation and metastasis. Cancer Metastasis Rev 2019;38:223-36. [Crossref] [PubMed]
- Gao XP, Dong JJ, Xie T, et al. Integrative Analysis of MUC4 to Prognosis and Immune Infiltration in Pan-Cancer: Friend or Foe? Front Cell Dev Biol 2021;9:695544. [Crossref] [PubMed]
- De Vita A, Vanni S, Fausti V, et al. Deciphering the Genomic Landscape and Pharmacological Profile of Uncommon Entities of Adult Rhabdomyosarcomas. Int J Mol Sci 2021;22:11564. [Crossref] [PubMed]
- Yang J, Yang D, Cogdell D, et al. APEX1 gene amplification and its protein overexpression in osteosarcoma: correlation with recurrence, metastasis, and survival. Technol Cancer Res Treat 2010;9:161-9. [Crossref] [PubMed]
- Teng S, Ma C, Yu Y, et al. Hydroxyurea promotes TET1 expression and induces apoptosis in osteosarcoma cells. Biosci Rep 2019;39:BSR20190456. [Crossref] [PubMed]
- Chiappetta C, Mancini M, Lessi F, et al. Whole-exome analysis in osteosarcoma to identify a personalized therapy. Oncotarget 2017;8:80416-28. [Crossref] [PubMed]
- Hong KH, Song S, Shin W, et al. A case of interdigitating dendritic cell sarcoma studied by whole-exome sequencing. Genes Genomics 2018;40:1279-85. [Crossref] [PubMed]
- Koba H, Kimura H, Nishikawa S, et al. Next-generation sequencing analysis identifies genomic alterations in pathological morphologies: A case of pulmonary carcinosarcoma harboring EGFR mutations. Lung Cancer 2018;122:146-50. [Crossref] [PubMed]
- Tamborero D, Gonzalez-Perez A, Perez-Llamas C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep 2013;3:2650. [Crossref] [PubMed]
- Sondka Z, Bamford S, Cole CG, et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 2018;18:696-705. [Crossref] [PubMed]
- Savas S, Skardasi G. The SWI/SNF complex subunit genes: Their functions, variations, and links to risk and survival outcomes in human cancers. Crit Rev Oncol Hematol 2018;123:114-31. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


