
Chromobox 4 (CBX4) promotes tumor progression and stemness via activating CDC20 in gastric cancer
Introduction
Gastric cancer (GC) a common malignancy of the stomach, and the third leading cause of cancer death worldwide. Based on the latest statistics, there would have been an estimated 26,560 new cases of GC, and the corresponding deaths would reach 11,180 in 2021 (1). As a complex disease that originates from the interplay between environment and host-related issues, the pathogenesis of GC is induced by many hazard factors, including Helicobacter pylori (Hp) infection, alcohol drinking, irregular diet, and genetics (2-4). The majority of GCs are adenocarcinomas, and can divided into intestinal and diffuse subtypes based on the Lauren classification system (5,6). Although traditional treatments comprise surgical resection and chemotherapy for patients with GC, their overall benefits have been limited (7). In recent decades, mounting attention has been directed towards the field of molecular targeted therapy (8-10). Intensive high-throughput sequencing technologies have deciphered the genomic profile of GC, finding novel molecular targets such as FZD7, EZH2, and ARID1A (11-13). In addition, The Cancer Genome Atlas (TCGA) database is a powerful resource for illustrating the molecular characterization of GC (14,15). Future challenges include the evaluation of clinical translational significance of these molecular targets, thereby providing valuable strategies for detection of GC and individualized management.
Epigenetic regulations maintain cell phenotypes or behaviors via mastering the transcriptional availability of vast regions across the genome, such as differential DNA methylation, chromatin accessibility, and DNA packaging via histone modifications (16,17). Cancer-associated epigenetic events exert effects via crosstalk or co-operation of mutually reinforcing or counteracting pathways (18,19). Genome-wide sequencing technologies have largely expanded the understanding of epigenetic markers and how they function (20). Recent studies on the aberrant modulation of gene promoters and enhancers in tumors have increased interest regarding the effects of epigenetic alterations on tumor formation (16,21). The epigenetic family members can be subdivided into transcription factors, histone writers, readers, and erasers. Overall, the recruitment, preservation, and modification of epigenetic marks are accurately modulated, with coordination among writers or marks to characterize the epigenetic profile (22,23). Polycomb group (PcG) proteins have been revealed as the key transcriptional repressors that promote epigenetic silencing of targets, including polycomb-repressive complexes (PRC) PRC1 and PRC2 (24). Particularly, polycomb chromobox (CBX) proteins participate in the PRC1 complex and grant the distinct biological roles for PRC1 (25-27). Meanwhile, CBX proteins may function as oncogenic drivers or tumor-suppressive factors in a cancer-type-dependent manner. It has been shown that CBX7 regulates the stemness traits of GC cells through p16 and AKT-NF-κB-miR-21 crosstalk (28). In contrast, CBX7 could also inhibit urinary bladder cancer progression through regulating the AKR1B10-ERK pathway (29). As a result, the specific roles of CBX proteins in each tumor should be evaluated and discussed separately. Among these members, chromobox homolog 4 (CBX4), also known as polycomb 2, is an unusual CBX protein that has dual functions to be both a SUMO E3 ligase and a transcriptional regulator (30,31). The two distinct functions of CBX4 are mainly mediated by the N-terminal chromodomain and two SUMO-interacting motifs (SIM). In recent years, CBX4 was identified as an effective vulnerability for hepatocellular carcinoma (HCC). Mechanistically, CBX4 could enhance HIF-1α sumoylation at K391 and K477 to increase the transcriptional activity of HIF-1. Meanwhile, the CBX4 proteins were positively associated with VEGF expressions, angiogenesis, and the overall survival (OS) of HCC patients (32). In addition, CBX4 recruits GCN5 to the Runx2 promoter to activate its levels, thereby enhancing osteosarcoma distal metastasis (33). However, no associated studies were available to discuss the relationships between CBX4 and GC tumorigenesis. Whether CBX4 is an oncogene or tumor suppressor in GC has yet to be determined.
In the current study, we used the bioinformatic analysis to identify that CBX4 is up-regulated in GC and is positively associated with poor survival outcomes. The growth and migration could be enhanced by CBX4. Based on these data, we provided novel insights into CBX4-mediated regulation of self-renewal capacity in GC. We also explored the epigenetic mechanism between CBX4 and CDC20, providing potential targets for GC treatment. We present the following article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-549/rc).
Methods
Cell culture and patient samples
We obtained GC cells (MGC-803, HGC-27, MKN-45, BGC-823) and 293 T cells from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China), and cultured the cells in Dulbecco’s Modified Eagle Medium (DMEM) medium, supplemented with 10% fetal bovine serum (FBS; Invitrogen, Camarillo, CA, USA). All cells were maintained at 37 ℃ in an atmosphere of 5% CO2.
Collection of GC samples and immunohistochemistry
All GC specimens were collected from The Second Affiliated Hospital, Hengyang Medical School, University of South China, and confirmed by pathology. For the IHC assay, GC tissues from patients were taken to paraffin imbedding and cut, and stained by hematoxylin. The 4 µm thick sections of tissues were sliced. After deparaffinization and rehydration, samples were blocked from endogenous peroxidases with 3% solution of hydrogen peroxide. Following this, IHC staining was performed using the specific primary antibodies against CBX4 according to standard protocols. After 1× PBS rinses for 15 min, tissue sections were incubated with the rabbit anti-goat biotinylated secondary antibody, and then followed by incubation with streptavidin-horseradish peroxidase complex (SABC) and stained with 3,3'-diaminobenzidine tetrahydrochloride hydrate (DAB). Sections were counterstained with hematoxylin. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of The Second Affiliated Hospital, Hengyang Medical School, University of South China, and informed consent was taken from all the patients.
Generation of CBX4-deleted GC cells
We selected MGC-803 and HGC-27 cells to construct CBX4-knockout (KO) cells, respectively. Briefly, the pX459 plasmid was utilized to clone sgRNA sequences that target CBX4. Then, MGC-803 and HGC-27 cells were planted into a 6-well plate and transfected with the constructed sequences. After one day, the puromycin (1 µg/mL) was used to screen GC cells for 72 hours. After drug screening, the remaining alive cells were further planted into a 96-well plate with the diluted concentration. The CBX4-KO cell clones were confirmed by western blot assay.
Colony formation assay and Transwell assay
For the colony formation assay, GC cell lines with CBX4 overexpression or CBX4-KO were diluted into a single-cell suspension. The 2,000 cells in each well of the 6-well plates were cultured under 5% CO2 at 37 ℃ for 10 days. After the cells were stained with 0.04% crystal violet and 2% ethanol, the colonies numbers were quantified and compared. The migration assay of BGC-823 and MGC-803 cells was performed in a 12-well Transwell plate with 8-mm polyethylene terephthalate membrane filters (Corning, Corning, NY, USA).
Tumorsphere formation assay
The ultra-low attachment plates (cat. no. 174925) were obtained from Corning Incorporated (Corning, USA). For the sphere formation assay, GC cells were plated in ultra-low attachment 96-well plates. Then, the cells were maintained in DMEM supplemented with 10% FBS. Afterwards, the sphere formation graphs were obtained after 14 days. We used 1,000 MGC-803 or HGC-27 cells for the sphere formation assay.
Western blotting assay
Cell proteins derived from the lysed cells were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which were further transferred to nitrocellulose membranes. Then, 5% milk was mixed with tris-buffered saline with Tween 20 [TBST; 120 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.05% Tween 20] to block the non-specific binding regions for 1 hour. After washing three times in phosphate-buffered saline (PBS), the membranes were further incubated with the primary antibodies of anti-CBX4 (Abcam, Cambridge, MA, USA; ab242149) and anti-CDC20 (Abcam, ab183479) overnight at 4 ℃. β-actin was used as a loading control. The next day, the membranes were washed with PBS three times. Then, the membranes were incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody. All indicated proteins were visualized at the bands using a Dura SuperSignal Substrate (Pierce, Waltham, MA, USA).
Chromatin immunoprecipitation (ChIP) assay
The MKN-45 cells were cross-linked at 37 ℃ for 10 minutes with the 1% formaldehyde. Then, 300 µL of lysis buffer was utilized to resuspend the cells. The DNA was divided by sonication method into several fragments, which were further diluted to a SDS mixture with 0.1% concentration. Using the herring sperm DNA and Protein A/G Magnetic beads (Thermo Fisher, Waltham, MA, USA), the recovered supernatants were incubated by specific antibodies or an isotype control immunoglobulin G (IgG) for 2 hours. The antibodies are the anti-CBX4 (Abcam, ab242149) and anti-H3K4me3 (Abcam, ab213224). The immunoprecipitated DNA was retrieved from the beads with 1% SDS and a 1.1MNaHCO3 solution, which was further purified by a polymerase chain reaction (PCR) Purification Kit (QIAGEN, Valencia, CA, USA).
Animal assay
For the in vivo metastatic model, MGC-803 cells were injected into nude mice via the tail vein portal (n=5/group). Bioluminescent (BIL) flux was used to detect the distal lung metastases with the unit of (photons/s/cm2/steradian). The IVIS-100 system (Caliper Life Sciences, Hopkinton, MA, USA) was used to monitor and evaluate the metastatic progression. After 6 weeks, all mice were euthanized. For the subcutaneous tumor model, 4–6-week-old BALB/c nu/nu mice were obtained from Shanghai Laboratory Animal Center (SLAC) Co., Ltd. (Shanghai, China). All mice were bred and maintained in our institutional pathogen-free mouse facilities. The 4×106 indicated BGC-823 cells were suspended with the 100 µL PBS buffer. Then, they were injected into the flanks of nude mice (n=5/group). At the end of 3 weeks, all mice were killed and the subcutaneous solid tumors were obtained and compared. Animal experiments were granted by ethics board of The Second Affiliated Hospital, Hengyang Medical School, University of South China, in compliance with the institutional guidelines for the care and use of animals.
Bioinformatic analysis
The expression matrix of CBX4 derived from GC patients was obtained from the TCGA resource via the Genomic Data Commons (GDC) portal (https://portal.gdc.cancer.gov/). We utilized the limma package to analyze the levels of CBX4 in tumor and normal tissues. Besides, we also downloaded the CBX4 expression data from GSE29272 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29272) with 134 patients, and GSE63089 (https://www.ncbi.nlm.nih.gov/gds/?term=GSE63089) with 45 paired samples, and GSE13195 (https://www.ncbi.nlm.nih.gov/gds/?term=GSE13195) with 50 paired samples. Boxplot was used to show the results. Kaplan-Meier analysis was carried out by the survival package. Gene set enrichment analysis (GSEA) was conducted using the CBX4 levels as the classification index. Based on the GSEA software running on Java platform, we obtained the “c2.cp.kegg.v6.2.symbols.gmt gene sets” from the Molecular Signatures Database (MSigDB; http://software.broadinstitute.org/gsea/msigdb) as the reference set. The enriched biological items with false discovery rate (FDR) <0.05 were considered statistically significant.
Statistical analysis
All data were expressed as mean values ± SD for experiments. Student’s t-test and one-way analysis of variance (ANOVA) were utilized to compare differences in continuous variables. Kaplan-Meier analysis with log-rank test was used to assess the prognostic value of CBX4 in GC samples. All statistical analyses were performed by using the software SPSS 19.0 (IBM Corp., Chicago, IL, USA). A P value <0.05 was considered statistically significant.
Results
Screening of CBX family members to identify CBX4 as a prognostic regulator in GC
To find the potential epigenetic regulators that are required for GC growth, we designed the specific small interfering RNAs (siRNAs) targeting CBX members in MGC-803 cells, individually. The 3-(4,5-dimethylthiazol-2-yl) (MTT) assay revealed that CBX4 inhibition induced the greatest decrease of cell growth relative to other family members (Figure 1A). Few studies have reported about the associations between CBX4 and GC. Besides, we carried out an analysis of Gene Expression Omnibus (GEO) databases incorporating GSE29272, GSE63089, and GSE13195 and discovered that messenger RNA (mRNA) levels of CBX4 were notably increased compared with those in the adjacent normal tissues (Figure 1B-1D). In addition, we also downloaded the transcriptome matrix of GC patients from TCGA dataset (https://portal.gdc.cancer.gov/). Differential analysis was conducted by limma package, which revealed that CBX4 was also higher in tumor than normal samples (N=408, P<0.001, Figure 1E). To assess whether CBX4 levels were related to prognosis of GC patients, we conducted Kaplan-Meier analysis based on the Kaplan-Meier plotter database (http://kmplot.com/analysis/index.php?p=service&cancer=gastric) to determine the effects of CBX4 on the survival of 875 samples. The prognostic analysis with survival curves revealed that patients with high CBX4 levels had shorter OS months or post-progression survival (PPS) months compared with those with low CBX4 levels (Figure 1F,1G). In summary, our screening data and bioinformatic analysis indicated that CBX4 is a hazard epigenetic regulator that has clinical prognostic significance in GC.

CBX4 promotes GC cells proliferation and growth
To further confirm the bioinformatic findings in the public datasets, we collected several GC samples with matched normal tissues to perform the immunohistochemistry (IHC) assay. In line with the above results and speculations, IHC staining indicated the overexpression of CBX4 in GC samples (Figure 2A). To determine the biological roles of CBX4 in GC, we selected two GC cell lines (MGC-803 and HGC-27) to establish CBX4-overexpressing cells. The protein and relevant mRNA levels of overexpressed CBX4 in MGC-803 and HGC-27 were confirmed by western blotting and quantitative (q)PCR assays (Figure 2B,2C). Overexpression of CBX4 enhanced the cell growth of MGC-803 and HGC-27 cells, indicating that CBX4 is an oncogenic factor in GC (Figure 2D). To further confirm these findings in vitro, we also used to CRISPR/Cas9 strategy to delete CBX4 in MKN-45 and BGC-823 cells (Figure 2E). As expected, CBX4 depletion significantly attenuated the cell proliferation ability of MKN-45 and BGC-823 cells relative to parental control cells (Figure 2F). In addition, CBX4 deficiency suppressed the colony formation abilities, whereas ectopic expressions of CBX4 could rescue the impaired abilities of MKN-45 cells (Figure 2G). In summary, these findings implied the oncogenic role of CBX4 in GC, showing that CBX4 could promote GC proliferation.
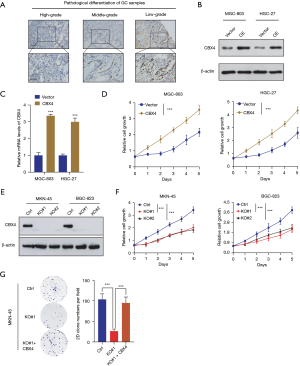
CBX4 enhances GC cells migration and invasion in vitro and in vivo
To further evaluate whether CBX4-mediated effects were required for GC metastasis, we assessed the relationships between CBX4 and GC cell motility. Firstly, CBX4 depletion notably attenuated the migratory ability of BGC823 cells as compared to WT control cells, as evidenced by the wound-healing assay (Figure 3A). Besides, ectopic expression of CBX4 significantly elevated the migration efficacy of MGC-803 cells (Figure 3B). In addition, Transwell Matrigel invasion assay further indicated that CBX4 KO remarkably impaired the invasive ability of BGC-823 cells (Figure 3C). However, CBX4 overexpression notably enhanced the invasive efficiency of MGC-803 cells (Figure 3D). Lastly, we also intended to determine the physiological roles of CBX4 on GC metastasis in vivo. The MGC-803 cells with modified CBX4 were stably established and subsequently injected into the BABL/c nude mice via the tail vein. The metastatic luciferase signals were detected at the indicated timepoints to monitor the location and growth of MGC-803-derived metastases. Apparently, we found that overexpression of CBX4 remarkably promoted the lung metastases lesions of MGC-803 cells relative to those in the control group, as detected by the bioluminescence signals in lung and numbers of metastatic nodes (Figure 3E,3F). In summary, these data implicated that CBX4 could facilitate GC cell migration, invasion, and in vivo metastasis.
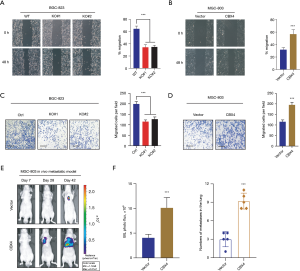
CBX4 regulated downstream CDC20 expressions to maintain cell stemness potentiality and growth
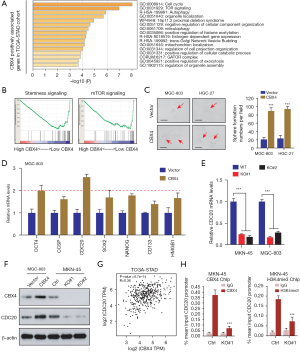
Considering the oncogenic roles of CBX4 in GC proliferation and metastasis, we questioned the potential crosstalk that CBX4 regulates in GC tumorigenesis. Firstly, we calculated the coefficients and obtained the top 353 genes with the highest CBX4 correlation coefficient based on the transcriptome data of the TCGA-stomach adenocarcinoma (STAD) cohort with Pearson’s r>0.3 (Table S1). Gene Ontology (GO) analysis was conducted to investigate the significantly enriched crosstalk and biological items based on these top genes. The results implicated that cell cycle, mTOR, and autophagy were the most enriched items, supporting the oncogenic roles of CBX4 in GC (Figure 4A). We then divided the TCGA-STAD samples into CBX4-high and CBX4-low groups using the median data of CBX4 as the cutoff. We conducted GSEA was carried out using the CBX4 levels (high vs. low) as the phenotype and we observed that stemness signaling was notably down-regulated in CBX-low samples (Figure 4B). Accordingly, we performed the tumorsphere formation assay to validate the bioinformatic findings, where CBX4 overexpression could notably enhance the self-renewal ability of GC cells (Figure 4C). Given that cancer stemness traits contribute to progression, metastasis, or drug resistance of GC, we wondered whether CBX4 could regulate the stemness-associated targets to modulate GC progression. We selected Ctrl and CBX4-overexpressing MGC-803 cells to screen the levels of representative stemness-associated genes, including OCT4, CCSP, CDC20, SOX2, and NANOG, among others. Compared with other genes, CDC20 mRNA levels showed the highest increase in response to CBX4 overexpression in MGC-803 cells (Figure 4D). In contrast, CBX4 KO could notably reduce the CDC20 mRNA levels in GC cells as compared to parental control cells (Figure 4E). We further conducted the western blotting assay to confirm that CBX4 could elevate CDC20 proteins, whereas CBX4 deletion caused the opposite effects (Figure 4F). Meanwhile, we also queried the transcriptome data of TCGA-STAD samples and found positive correlations between CBX4 and CDC20 TPM levels with Pearson r=0.36 (Figure 4G). Previous studies have already indicated CBX proteins are responsible for recognizing histone 3 lysine trimethylation at residues K9 and K27 to mediate epigenetic modifications (34,35). As reported, CBX4 could coordinate with the histone H3 protein subunit (H3K4me3), a modification commonly associated with active transcription, at the TOP2α promoter to activate its expressions in LoVo cells (36). We thus conducted the ChIP-qPCR to confirm that CBX4 and H3K4me3 co-occupied on the promoter region of CDC20 (Figure 4H). Meanwhile, CBX4 ablation resulted in an 85% decrease of CBX4 enrichment at the CDC20 promoter of MKN-45 cells. We also found a decrease in H3K4me3 abundance at the promoter loci of CDC20 in CBX4-deficient cells (Figure 4H). Collectively, these data indicated that CBX4 mainly accompanies with H3K4me3 to activate CDC20 levels, thereby enhancing self-renewal capacity and progression of GC.
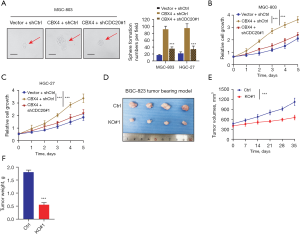
Targeting CBX4 is effective to suppress GC progression
To further elucidate the clinical relevance of CBX4/CDC20 axis in regulating GC tumorigenesis, we conducted the short hairpin RNA (shRNA)-mediated knockdown assay to target CDC20 via lentiviruses in CBX4-overexpressing cells. Notably, CBX4 could enlarge the sphere sizes, while CDC20 inhibition restored the CBX4-mediated effect (Figure 5A). Then, Cell Counting Kit-8 (CCK-8) assays were simultaneously conducted to find that CBX4 relied on CDC20 to potentiate in vitro GC cell growth (Figure 5B,5C). Lastly, stable parental and CBX4-deficient BGC-823 cells were implanted into nude mice and the xenograft tumor volumes were detected every seven days. In line with expectations, CBX4 deficiency significantly inhibited tumor growth, exhibiting significantly smaller tumor sizes and tumor weight (Figure 5D-5F). Taken together, these data suggested that targeting CBX4 is an effective strategy to inhibit GC progression, creating an epigenetic vulnerability for clinical utility.
Discussion
Currently, GC is a common gastrointestinal tumor worldwide. Apart from the genetic or environmental impacts, epigenetic regulations are still the main reason in the origination of GC. As is well documented, the epigenetic alterations mainly incorporate DNA methylation, post-translational modifications, and chromatin remodeling events that participate in the progression of tumors. Among the other epigenetic alterations, DNA methylation occupies a special space (37-39). The methylation or de-methylation of DNA is regulated by various tumor suppressors or oncogenes that participate in multiple steps of tumorigenesis, like cell adhesion, cell proliferation, invasion, DNA repair signaling (40,41). The epigenetic modifications in modulating expressions of many cancer-related genes have played essential roles in GC initiation and progression and formed novel epigenetic signature (42-44). Targeting key epigenetic drivers would notably suppress cancer progression for GC (45). As is well known, PRC1 and PRC2 co-operate to regulate epigenetics via histone modification incorporating the methylation of histone H3K27 and monoubiquitination of histone H2AK119 (46,47). As the vital component of PRC1, CBX4 was shown to be an essential regulator that mediates the epigenetic modifications in various tumors (26,48,49). In the current study, we comprehensively screened the polycomb CBX members using siRNAs in GC cells. Intriguingly, CBX4 inhibition induced the greatest decrease of cell growth. Subsequent bioinformatic analysis further indicated that CBX4 was upregulated in multiple GC datasets and patients with high CBX4 correlated with poorer prognosis relative to those with low CBX. CBX4 could be used as a potential biomarker for GC patient risk stratification and local regional metastasis. Enforced expression of CBX4 enhanced the cell growth, whereas CBX4 deficiency inhibited GC proliferation. Besides, CBX4 could also enhance the cell migration and invasive properties in vitro and in vivo. Mechanistically, GSEA revealed that CBX4 could modulate the self-renewal process, sustaining stemness features of GC. We further screened and validated that CDC20 was the downstream target of CBX4. Inhibition of CBX4 could reduce the CDC20 levels, and CBX4 could mediate the H3K4me3 enrichment at the promoter region of CDC20 to sustain its levels. Additionally, we also demonstrated that CBX4 could depend on CDC20 to drive cell growth and tumorsphere formation capacity. Targeting CBX4 induced the impaired in vivo GC growth, implicating the translational significance.
It was well documented that the GC stem-like cells (GCSCs) within the tumors contribute to the malignant features and progression such as drug resistance, distal metastasis, and recurrence (50,51). These CSCs have the potentialities to self-renew and differentiate into different cell subtypes. Several specific markers were identified in CSCs, which determine the subtype of tumors and characteristic subpopulation of cells. These markers contain CD133, CXCR4, LGR5, CD90, as well as CD44 (52,53). The original properties of primary tumors determine the levels of these markers. Therefore, it makes sense to conduct well-designed studies to comprehensively investigate the GCSC subpopulation within each tumor or cell line. Overall, CSCs could mainly activate the downstream signaling, including Notch, Wnt, β-catenin, and epithelial to mesenchymal transition (EMT). Wu et al. found that mesenchymal stem cell (MSC)-induced long non-coding RNA (lncRNA) HCP5 promoted fatty acid oxidation (FAO) via the miR-3619-5p/AMPK/PGC1α/CEBPB axis to modulate stemness and chemo-resistance of GC, suggesting that HCP5 inhibition is effective to synergize the efficacy of chemotherapy in GC (54). In addition, MiR-375 could regulate the downstream target SLC7A11 to modulate ferroptosis in GC, which further impacts the stemness maintenance (55). In the current study, we found that CBX4 could regulate the GC stemness via activating CDC20 expressions. Zhang et al. have revealed that CDC20 sustains the stemness property of CD44+ prostate CSCs by enhancing the nuclear translocation and trans-activation of β-catenin. Therefore, CDC20 could be utilized to be the effective predictors for the prognosis of prostate cancer patients, along with CD44 or β-catenin (56). Besides, researchers have conducted weighted gene co-expression network analysis (WGCNA) to analyze the stemness-related genes (SRGs) and CDC20 has been shown to be among these hub genes, correlating with stem-like characteristics of tumor microenvironment (TME) of lung cancer (57). Considering that CBX4 depends on CDC20 to drive stemness of GC, we speculated that CDC20 inhibition could effectively suppress the stemness and proliferation of GC, highlighting a novel strategy for treatment.
However, there were several limitations to the current study. Firstly, we need to collect more GC samples in our hospital with complete clinical survival information to assess the relationships between CBX4 and GC prognosis. Secondly, we need to construct more pre-clinical models to assess the biological relationships between CBX4 and GC progression, including patient-derived xenografts (PDXs), patient-derived organoids (PDOs), and orthotopic GC tumor model. In addition, we conducted the GSEA to find several oncogenic pathways that were related to CBX4. Apart from stemness, whether CBX4 regulated other signaling, like cell cycle, mTOR, or VEGF, remains to be investigated further. Last of all, further research should investigate the potential up-stream mechanisms that contribute to high CBX4 levels in GC.
Conclusions
Taken together, our study revealed the clinical significance of CBX4 in GC. High CBX4 correlated with poor prognosis and advanced clinical characteristics. High CBX4 enhanced tumor proliferation, migration, and stemness in vitro and in vivo. It was revealed that CBX4 coordinated with H3K4me3 to maintain CDC20 levels, thereby elevating cancer stemness capacities. Targeting the CBX4/CDC20 axis creates an epigenetic vulnerability in GC treatment.
Acknowledgments
Funding: This study was supported by the Natural Science Foundation of Hunan, China (No. 2020JJ4537).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-549/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-549/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-549/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. For research involving human, the study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of The Second Affiliated Hospital, Hengyang Medical School, University of South China, and informed consent was taken from all the patients. Animal experiments were granted by ethics board of The Second Affiliated Hospital, Hengyang Medical School, University of South China, in compliance with the institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Correa P. Gastric cancer: overview. Gastroenterol Clin North Am 2013;42:211-7. [Crossref] [PubMed]
- Alipour M. Molecular Mechanism of Helicobacter pylori-Induced Gastric Cancer. J Gastrointest Cancer 2021;52:23-30. [Crossref] [PubMed]
- Pilonis ND, Tischkowitz M, Fitzgerald RC, et al. Hereditary Diffuse Gastric Cancer: Approaches to Screening, Surveillance, and Treatment. Annu Rev Med 2021;72:263-80. [Crossref] [PubMed]
- Pernot S, Terme M, Radosevic-Robin N, et al. Infiltrating and peripheral immune cell analysis in advanced gastric cancer according to the Lauren classification and its prognostic significance. Gastric Cancer 2020;23:73-81. [Crossref] [PubMed]
- Yin X, Fang T, Wang Y, et al. Prognostic significance of serum inflammation indexes in different Lauren classification of gastric cancer. Cancer Med 2021;10:1103-19. [Crossref] [PubMed]
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635-48. [Crossref] [PubMed]
- Lee J, Kim ST, Kim K, et al. Tumor Genomic Profiling Guides Patients with Metastatic Gastric Cancer to Targeted Treatment: The VIKTORY Umbrella Trial. Cancer Discov 2019;9:1388-405. [Crossref] [PubMed]
- Singh P, Toom S, Huang Y. Anti-claudin 18.2 antibody as new targeted therapy for advanced gastric cancer. J Hematol Oncol 2017;10:105. [Crossref] [PubMed]
- Meric-Bernstam F, Johnson AM, Dumbrava EEI, et al. Advances in HER2-Targeted Therapy: Novel Agents and Opportunities Beyond Breast and Gastric Cancer. Clin Cancer Res 2019;25:2033-41. [Crossref] [PubMed]
- Pi J, Wang W, Ji M, et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res 2021;81:2651-65. [Crossref] [PubMed]
- Han H, Wang S, Meng J, et al. Long noncoding RNA PART1 restrains aggressive gastric cancer through the epigenetic silencing of PDGFB via the PLZF-mediated recruitment of EZH2. Oncogene 2020;39:6513-28. [Crossref] [PubMed]
- Lo YH, Kolahi KS, Du Y, et al. A CRISPR/Cas9-Engineered ARID1A-Deficient Human Gastric Cancer Organoid Model Reveals Essential and Nonessential Modes of Oncogenic Transformation. Cancer Discov 2021;11:1562-81. [Crossref] [PubMed]
- Chen S, Wei Y, Liu H, et al. Analysis of Collagen type X alpha 1 (COL10A1) expression and prognostic significance in gastric cancer based on bioinformatics. Bioengineered 2021;12:127-37. [Crossref] [PubMed]
- Li X, Pasche B, Zhang W, et al. Association of MUC16 Mutation With Tumor Mutation Load and Outcomes in Patients With Gastric Cancer. JAMA Oncol 2018;4:1691-8. [Crossref] [PubMed]
- Hogg SJ, Beavis PA, Dawson MA, et al. Targeting the epigenetic regulation of antitumour immunity. Nat Rev Drug Discov 2020;19:776-800. [Crossref] [PubMed]
- Costa-Pinheiro P, Montezuma D, Henrique R, et al. Diagnostic and prognostic epigenetic biomarkers in cancer. Epigenomics 2015;7:1003-15. [Crossref] [PubMed]
- Chiappinelli KB, Zahnow CA, Ahuja N, et al. Combining Epigenetic and Immunotherapy to Combat Cancer. Cancer Res 2016;76:1683-9. [Crossref] [PubMed]
- Zhang C, Lu X, Huang J, et al. Epigenome screening highlights that JMJD6 confers an epigenetic vulnerability and mediates sunitinib sensitivity in renal cell carcinoma. Clin Transl Med 2021;11:e328. [Crossref] [PubMed]
- Zhang C, Chen L, Lou W, et al. Aberrant activation of m6A demethylase FTO renders HIF2αlow/- clear cell renal cell carcinoma sensitive to BRD9 inhibitors. Sci Transl Med 2021;13:eabf6045. [Crossref] [PubMed]
- Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, et al. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics 2019;14:1164-76. [Crossref] [PubMed]
- Skvortsova K, Iovino N, Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol 2018;19:774-90. [Crossref] [PubMed]
- Orozco-Solis R, Aguilar-Arnal L. Circadian Regulation of Immunity Through Epigenetic Mechanisms. Front Cell Infect Microbiol 2020;10:96. [Crossref] [PubMed]
- Blackledge NP, Farcas AM, Kondo T, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014;157:1445-59. [Crossref] [PubMed]
- Sun D, Cao X, Wang C. Polycomb chromobox Cbx2 enhances antiviral innate immunity by promoting Jmjd3-mediated demethylation of H3K27 at the Ifnb promoter. Protein Cell 2019;10:285-94. [Crossref] [PubMed]
- Wang Z, Fang Z, Chen G, et al. Chromobox 4 facilitates tumorigenesis of lung adenocarcinoma through the Wnt/β-catenin pathway. Neoplasia 2021;23:222-33. [Crossref] [PubMed]
- Klauke K, Radulović V, Broekhuis M, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol 2013;15:353-62. [Crossref] [PubMed]
- Ni SJ, Zhao LQ, Wang XF, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J Hematol Oncol 2018;11:17. [Crossref] [PubMed]
- Huang Z, Yan Y, Zhu Z, et al. CBX7 suppresses urinary bladder cancer progression via modulating AKR1B10-ERK signaling. Cell Death Dis 2021;12:537. [Crossref] [PubMed]
- Sanyal S, Mondal P, Sen S, et al. SUMO E3 ligase CBX4 regulates hTERT-mediated transcription of CDH1 and promotes breast cancer cell migration and invasion. Biochem J 2020;477:3803-18. [Crossref] [PubMed]
- Pan Y, Li Q, Cao Z, et al. The SUMO E3 ligase CBX4 is identified as a poor prognostic marker of gastric cancer through multipronged OMIC analyses. Genes Dis 2021;8:827-37. [Crossref] [PubMed]
- Li J, Xu Y, Long XD, et al. Cbx4 governs HIF-1α to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell 2014;25:118-31. [Crossref] [PubMed]
- Wang X, Qin G, Liang X, et al. Targeting the CK1α/CBX4 axis for metastasis in osteosarcoma. Nat Commun 2020;11:1141. [Crossref] [PubMed]
- van Wijnen AJ, Bagheri L, Badreldin AA, et al. Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development. Bone 2021;143:115659. [Crossref] [PubMed]
- Jung J, Buisman SC, Weersing E, et al. CBX7 Induces Self-Renewal of Human Normal and Malignant Hematopoietic Stem and Progenitor Cells by Canonical and Non-canonical Interactions. Cell Rep 2019;26:1906-1918.e8. [Crossref] [PubMed]
- Silva-Fisher JM, Dang HX, White NM, et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat Commun 2020;11:2156. [Crossref] [PubMed]
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27-56. [Crossref] [PubMed]
- Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 2018;19:371-84. [Crossref] [PubMed]
- Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol 2018;15:459-66. [Crossref] [PubMed]
- Lau CE, Robinson O. DNA methylation age as a biomarker for cancer. Int J Cancer 2021;148:2652-63. [Crossref] [PubMed]
- Wang M, Ngo V, Wang W. Deciphering the genetic code of DNA methylation. Brief Bioinform 2021;22:bbaa424. [Crossref] [PubMed]
- Ramezankhani R, Solhi R, Es HA, et al. Novel molecular targets in gastric adenocarcinoma. Pharmacol Ther 2021;220:107714. [Crossref] [PubMed]
- Huo FC, Zhu ZM, Zhu WT, et al. METTL3-mediated m6A methylation of SPHK2 promotes gastric cancer progression by targeting KLF2. Oncogene 2021;40:2968-81. [Crossref] [PubMed]
- Woo Y, Behrendt CE, Yang A, et al. Tumor Epigenetic Signature and Survival in Resected Gastric Cancer Patients. J Am Coll Surg 2021;232:483-491.e1. [Crossref] [PubMed]
- Canale M, Casadei-Gardini A, Ulivi P, et al. Epigenetic Mechanisms in Gastric Cancer: Potential New Therapeutic Opportunities. Int J Mol Sci 2020;21:5500. [Crossref] [PubMed]
- Zepeda-Martinez JA, Pribitzer C, Wang J, et al. Parallel PRC2/cPRC1 and vPRC1 pathways silence lineage-specific genes and maintain self-renewal in mouse embryonic stem cells. Sci Adv 2020;6:eaax5692. [Crossref] [PubMed]
- Healy E, Mucha M, Glancy E, et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol Cell 2019;76:437-452.e6. [Crossref] [PubMed]
- Zhao W, Ma B, Tian Z, et al. Inhibiting CBX4 efficiently protects hepatocellular carcinoma cells against sorafenib resistance. Br J Cancer 2021;124:1237-48. [Crossref] [PubMed]
- Jiao HK, Xu Y, Li J, et al. Prognostic significance of Cbx4 expression and its beneficial effect for transarterial chemoembolization in hepatocellular carcinoma. Cell Death Dis 2015;6:e1689. [Crossref] [PubMed]
- Zhang ZZ, Yu WX, Zheng M, et al. PIN1 Inhibition Sensitizes Chemotherapy in Gastric Cancer Cells by Targeting Stem Cell-like Traits and Multiple Biomarkers. Mol Cancer Ther 2020;19:906-19. [Crossref] [PubMed]
- Yoon C, Lu J, Yi BC, et al. PI3K/Akt pathway and Nanog maintain cancer stem cells in sarcomas. Oncogenesis 2021;10:12. [Crossref] [PubMed]
- Leon G, MacDonagh L, Finn SP, et al. Cancer stem cells in drug resistant lung cancer: Targeting cell surface markers and signaling pathways. Pharmacol Ther 2016;158:71-90. [Crossref] [PubMed]
- Azimi A, Mohaqiq M, Movahedin M, et al. Characterization of embryonic stem-like cells derived from mouse spermatogonial stem cells following low-intensity ultrasound treatment. Rev Int Androl 2021;19:264-71. [PubMed]
- Wu H, Liu B, Chen Z, et al. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1α/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis 2020;11:233. [Crossref] [PubMed]
- Ni H, Qin H, Sun C, et al. MiR-375 reduces the stemness of gastric cancer cells through triggering ferroptosis. Stem Cell Res Ther 2021;12:325. [Crossref] [PubMed]
- Zhang Q, Huang H, Liu A, et al. Cell division cycle 20 (CDC20) drives prostate cancer progression via stabilization of β-catenin in cancer stem-like cells. EBioMedicine 2019;42:397-407. [Crossref] [PubMed]
- Zeng H, Ji J, Song X, et al. Stemness Related Genes Revealed by Network Analysis Associated With Tumor Immune Microenvironment and the Clinical Outcome in Lung Adenocarcinoma. Front Genet 2020;11:549213. [Crossref] [PubMed]
(English Language Editor: J. Jones)


