
TRIM29 is differentially expressed in colorectal cancers of different primary locations and affects survival by regulating tumor immunity based on retrospective study and bioinformatics analysis
Introduction
Colorectal cancer (CRC) ranks third in incidence and death rate among all malignant tumors (1). The location of CRC has become an important topic. Since proximal and distal CRC arises through different embryonic layers, Bufill et al. suggested that these are two distinct types of cancers (2). Furthermore, rectal cancer (RECC) differs from colon cancer in anatomy, aetiology, clinical manifestation, biological features, treatment response, and clinical outcomes (3,4). Therefore, dividing CRC into left colon cancer (LCC) and right colon cancer (RCC) may be overly simplistic and most specialists now classify CRC into LCC, RCC, and RECC. There are obvious differences in clinicopathologic features and survival depending on the location of the tumors. These differences were first observed between LCC and RCC in metastasis CRC patients (5). The differences in survival between patients with different primary tumor locations may be due to various sensitivities to target drugs, as suggested in the FIRE-3 and CALGB/SWOG 80405 trials (6). A study has also shown that primary tumor location may be predictive of early stage and locally advanced CRC (7). We hypothesize that the differences not only result from various sensitivities to target drugs, but may be due to the molecular mechanisms involved (8). Papagiorsis’ study showed that there were different chromosomal variations and epigenetic changes in LCC and RCC patients, with at least 1,000 genes in the major signaling pathways significantly differentially expressed (9). The key oncogenes and tumor suppressors were differentially expressed, including APC, TP53, KRAS, and EGFR (9-11). In recent years, novel biomarkers were found to be differentially expressed between LCC and RCC, including G protein subunit gamma 4 (GNG4), Transcobalamin 1 (TCN1), and dual-specificity phosphatase-2 (DUSP2) (12,13). However, to date, the driver gene and the mechanisms underlying the differences observed in patients with different tumor locations remain unclear. Our previous study identified an upregulated gene (TRIM29) in RCC patients compared to LCC patients (14). TRIM29 has verified as an oncogene in many kinds of cancers, and our previous study has confirmed it play an important role in the tumorigenesis and development of CRC. More interestingly, we also found it differently expressed between LCC and RCC. However, the sample size in the latter study was small and the expression of TRIM29 was not investigated in RECC patients. Furthermore, the effects of TRIM29 on the survival of patients with different tumor locations were not examined. This current study further confirmed that TRIM29 expression is highly expressed in proximal CRCs compared to distal CRCs by using a large dataset from the GEO database. The differing roles of TRIM29 were examined in patients with tumors in three different sites. More importantly, the potential molecular mechanisms of TRIM29 were investigated. Reactome/Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the hub genes related to TRIM29 was conducted using the String database. Combined with the KEGG pathway enrichment analyses in our previous study (15), we hypothesized that TRIM29 is significantly correlated to immune activity. Interestingly, it has been reported that the immune microenvironment is different in patients with different tumor locations (16,17). Thus, the correlation between TRIM29 expression and immune function was assessed using the Tumor-Immune System Interactions and Drug Bank database (TISIDB). The findings herein may reveal the potential molecular mechanisms of CRC according to tumor site. TRIM29 may be a prognostic marker of RCC and a predictive marker of immunotherapy. We present the following article in accordance with the REMARK reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-365/rc).
Methods
This study mainly includes four steps. Firstly, we proved TRIM29 were highly expressed in RCC patients. A GEO database was used to analyze TRIM29 expression in proximal and distal CRCs. Reverse transcription polymerase chain reaction (RT-PCR), IHC and Western blotting were used testing TRIM29 expression level to classify TRIM29 was significantly highly expressed in RCC patients. Secondly, a retrospective analysis of 227 patients was conducted to analyze the value of TRIM29 in RCC, LCC and RECC patients respectively. The relationship between TRIM29 and clinicopathological parameters was also explored. Kaplan-Meier analysis was used to discuss the relationship between TRIM29 expression/clinicopathological features and survival. Thirdly, Bioinformatics analysis were used to explore the potential molecular mechanisms and the results focused that TRIM29 may modulate tumor-associated immunity. At last, GEO database and TISIDB were used to explore the relationship between TRIM29 and tumor-associated immunity. H&E staining was applied to observe the tumor infiltrating lymphocytes
Access to public data
An expression profiling dataset [GSE39582(GPL570 platform)], containing 233 proximal colorectum samples and 350 distal colorectum samples, was obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo). The TRIM29 expression level, tumor location, and the mismatch-repair-proficient/mismatch-repair-deficient (pMMR/dMMR) status of each sample in the dataset was recorded and analyzed.
Patient characteristics
The medical records of 227 patients with CRC, who were treated at the Hebei Medical University Fourth Affiliated Hospital between January 2008 and January 2015, were retrospectively reviewed. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The collection of samples in this study was approved by ethics board of Hebei Medical University Fourth Affiliated Hospital (No. 2017MEc089). Informed consent was obtained from all patients. The following inclusion criteria were applied: (I) patients with pathologically confirmed CRC; and (II) patients who had undergone curative surgical resection (primary tumor and all metastases were excised together). The following patients were excluded: (I) patients with two or more primary tumors; (II) patients with adenocarcinoma and other types of malignancies affecting the appendix; (III) patients with tumors of unknown location; and (IV) patients with incomplete information. Disease staging was performed according to the seventh edition of the American Joint Committee on Cancer’s TNM classification. The patients’ demographic and clinicopathological characteristics were collated from a medical data platform by trained staff, who used standardized data collection and quality-control procedures. Patients were followed up by hospital visits for regular review or followed up by telephone visits every 2 months. The end of the follow-up time was January 2021, and the median follow-up time was 48.2 months. Disease-free survival (DFS) was defined as the time from surgery to the tumor recurrence and metastasis, overall survival (OS) was defined as the time from surgery to death of any cause or the loss of follow-up.
Among all patients enrolled, 35.24% (n=80) had RCC, 37.00% (n=84) presented with LCC, and 27.75% (n=63) had RECC. Among the RCC patients, 44 (55.00%) were men and 36 (45.00%) were women. There were 16 patients (20.00%) under the age of 60 years and 64 patients (80.00%) were 60 years or older. Among the LCC patients, 60 (71.43%) were male and 24 (28.57%) were female. There were 32 patients (38.10%) under the age of 60 years and 52 patients (61.90%) aged 60 years or older. Among the RECC patients, 25 (39.68%) were male and 38 (60.32%) were female. There were 39 (61.90%) patients aged 60 years and 24 (38.10%) patients aged 60 years or older.
Six clinicopathological features (gender, age, intestinal obstruction, TNM stage, T stage, N stage) are important in clinical treatment and may relate to prognosis. So the relationship between TRIM29 and these six clinical covariates was analyzed.
Fresh-frozen tissue samples from human CRC
A total of 68 fresh-frozen human CRC tissues were obtained from Hebei Medical University Fourth Affiliated Hospital, including 4 LCC tissues, 24 RCC samples, and 20 RECC tissues. All samples were pathologically confirmed as either colorectal adenocarcinoma tissues or normal colorectal tissues.
Paraffin sectioning and immunohistochemical staining
CRC tissues and matched normal tissues were obtained from the 227 patients. Paraffin sections were prepared and deparaffinized in xylene and dehydrated in a graded series of ethanol. Endogenous peroxidase activity was blocked using 3% H2O2 solution, followed by point antigen retrieval using a 20-minute heat-induced antigen retrieval procedure in pH 9.0 TRIS-EDTA buffer (zsbio). The slides were probed with an anti-TRIM29 antibody (Signalway Antibody). Protein expression was measured using a 3,3-diaminobenzidine (DAB) peroxidase substrate (zsbio).
Interpretation of the histology and immunohistochemical staining results
The IHC staining results of TRIM29 were independently evaluated by two pathologists who were not aware of the patients’ clinical outcomes. A consensus decision was made through consultation when they had an interobserver discrepancy. The staining intensity was scored according to the following criteria: 0, no staining; 1+, weak staining; 2+, moderate staining; and 3+, strong staining. High IHC expression was defined as a staining intensity of 3+ in over 25% of tumor cells.
Hematoxylin and eosin staining and tumor lymphocytic infiltration
Tissue specimens were stained with hematoxylin and eosin (H&E) and the tumor lymphocytic infiltration was evaluated. Tumor lymphocytic infiltration greater than 50% was defined as high tumor lymphocytic infiltration.
RNA extraction, RT-PCR, and quantitative real-time PCR (qPCR)
Total RNA was extracted from 68 tissue samples (including 20 RCC samples, 24 LCC samples, and 24 RECC samples) with TRIzol reagent (Invitrogen). The cDNA was synthesized with OneScript Plus Reverse Transcriptase (abm). The qPCR was conducted with the OneScript Plus cDNA Synthesis kit (abm). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal normalization reference.
The qPCR primers used are listed as follows:
TRIM29 forward: AGCATCAGCGACTCTGTGTTG;
TRIM29 reverse: GAAGTTGCCTAGTGACTGTCC;
GAPDH forward: GAGAAGGCTGGGGCTCATTT;
GAPDH reverse: AGTGATGGCATGGACTGTGG.
Western blot analysis
All fresh-frozen tumor samples were lysed using RIPA lysis buffer (Applygen, China) containing a protease inhibitor cocktail. Thereafter, 30 µg of protein lysate per sample was separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a 10% gel and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% skim milk for at least 1 hour at 37 ℃, followed by incubation with primary antibodies overnight at 4 ℃. The primary antibodies employed were anti-TRIM29 (Signalway Antibody) and anti-β-actin (Santa Cruz Biotechnology). Then, the membranes were probed with the corresponding secondary antibodies (Promega, USA) and incubated with a chemiluminescent substrate to form the protein bands. Images were taken using an Image Reader LAS-4000 (Fuji Ltd., 120 Japan).
Construction of a protein-protein interaction (PPI) network and determining the functional annotations of TRIM29
The Search Tool for the String application (http://string.embl.de/), an open-source online tool, was used to identify the proteins which closely correlated with TRIM29 and a PPI network was constructed. A local clustering coefficient greater than 0.875 was considered as significantly related hub genes. Subsequently, gene ontology (GO) analysis, including biological processes (BP), molecular function (MF), and cellular component (CC), and Reactome/KEGG pathways analysis were performed for the hub genes associated with TRIM29.
Construction of the relationship between TRIM29 and immune infiltration
An integrated repository portal for tumor-immune system interactions (TISIDB, http://cis.hku.hk/TISIDB/) was used to examine the correlation between TRIM29 expression and tumor infiltrating lymphocytes, as well as immunomodulators and chemokines.
Statistical analyses
All statistical analyses were performed using the IBM SPSS Statistics ver. 22.0 (IBM Co., Armonk, NY, USA). Figures were prepared using GraphPad Prism 5 software. Categorical variables were analyzed using the chi-square test. Continuous variables were analyzed using the Student’s t-test. DFS and OS was analyzed using the Kaplan-Meier method, and comparisons were performed using the log-rank test. The Cox proportional-hazards model was performed for multivariate analyses to identify the prognostic factors for DFS and OS. The other clinical covariates were corrected as confounding factors by using Cox proportional-hazards model, when analyzing the independent prognostic role of TRIM29. Spearman’s test was used to measure correlations between TRIM29 and immune functions. All statistical tests were two-sided, and a P value less than 0.05 was considered statistically significant.
Results
TRIM29 is highly expressed in proximal CRC patients compared to distal CRC patients
The GSE39582 dataset [GPL570(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array] was downloaded from the GEO database. A total of 583 samples, consisting of 233 proximal colorectum samples and 350 distal colorectum samples, were analyzed for TRIM29 expression levels. The results showed that proximal colorectum cancer patients expressed higher TRIM29 levels than distal CRC patients (see Figure 1A, P=0.0006).
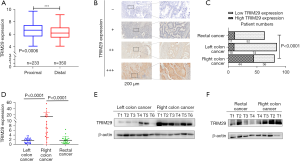
TRIM29 expression is significantly higher in RCC than in LCC and RECC
To further determine the TRIM29 expression in tumors at different locations, qPCR, Western blotting, and IHC was used to detect the TRIM29 mRNA and protein levels.
IHC was used to detect TRIM29 protein levels in 227 paraffin sections of CRC. Among them, 80 of the patients had RCC, 84 patients presented with LCC, and 63 had RECC. The results revealed that TRIM29 protein expression was significantly higher in RCC samples than in LCC and RECC samples (Figure 1B,1C, P<0.001).
The qPCR was used to detect the TRIM29 mRNA level in a cohort of 68 CRC tissues, including 20 RCC, 24 LCC, and 24 RECC samples. The qPCR results demonstrated that TRIM29 mRNA expression was significantly higher in RCC compared to LCC and RECC (Figure 1D, P<0.0001). Then, 16 of the 68 samples were subjected to Western blotting in which 6 RCC tumor tissues were compared with 6 LCC samples, and 4 of the 6 RCC tumor tissues were compared with 4 RECC tumor tissues. The results revealed that TRIM29 protein expression was significantly higher in RCC compared to LCC and RECC (Figure 1E,1F).
TRIM29 is associated with different clinicopathologic features in patients with different tumor sites
The relationship between the TRIM29 expression and clinicopathological parameters was analyzed separately in RCC, LCC, and RECC patients. Six main clinicopathological parameters may affect prognosis were included in the analysis. In LCC patients, high TRIM29 expression was associated with intestinal obstruction before surgery (P=0.006). In RECC patients, high TRIM29 expression was associated with more advanced stage and younger age (P=0.015 and P=0.000, respectively). In patients with RCC, high TRIM29 expression was associated with the male sex, older age, stage III–IV, N+ staging, and intestinal obstruction (P≤0.010) (Tables 1-3).
Table 1
| Factor | N | High TRIM29 expression (n=10) | Low TRIM29 expression (n=74) | χ2 | P value |
|---|---|---|---|---|---|
| Gender | 3.090 | 0.079 | |||
| Male | 60 | 10 (100%) | 50 (67.57%) | ||
| Female | 24 | 0 (0%) | 24 (32.43%) | ||
| Age (years) | 5.272 | 0.22 | |||
| <60 | 32 | 0 (0%) | 32 (43.24%) | ||
| ≥60 | 52 | 10 (100%) | 42 (56.76%) | ||
| Intestinal obstruction | 7.599 | 0.006 | |||
| No | 66 | 4 (40%) | 62 (83.78%) | ||
| Yes | 18 | 6 (60%) | 12 (16.22%) | ||
| TNM stage | 0.009 | 0.923 | |||
| Stage I–II | 45 | 6 (60%) | 39 (52.70%) | ||
| Stage III–IV | 39 | 4 (40%) | 35 (47.30%) | ||
| T stage (Serosa involved) | 2.637 | 0.104 | |||
| T1–T3 (no) | 22 | 0 (0%) | 22 (29.73%) | ||
| T4 (yes) | 62 | 10 (100%) | 52 (70.27%) | ||
| N stage | 0.454 | 0.500 | |||
| N0 | 42 | 6 (60%) | 36 (48.65%) | ||
| N+ | 42 | 4 (40%) | 38 (51.35%) |
Table 2
| Factor | N | High TRIM29 expression (n=11) | Low TRIM29 expression (n=52) | χ2 | P value |
|---|---|---|---|---|---|
| Gender | 0.008 | 0.927 | |||
| Male | 25 | 5 (45.45%) | 20 (38.46%) | ||
| Female | 38 | 6 (54.55%) | 32 (61.54%) | ||
| Age (years) | 16.405 | 0.000 | |||
| <60 | 39 | 11 (100%) | 28 (53.85%) | ||
| ≥60 | 24 | 0 (0%) | 24 (46.15%) | ||
| Intestinal obstruction | 1.818 | 0.178 | |||
| No | 51 | 11 (100%) | 40 (76.93%) | ||
| Yes | 12 | 0 (0%) | 12 (23.07%) | ||
| TNM stage | 5.950 | 0.015 | |||
| I–II | 49 | 5 (45.45%) | 44 (84.62%) | ||
| III–IV | 14 | 6 (54.55%) | 8 (15.38%) | ||
| T stage (Serosa involved) | 1.818 | 0.178 | |||
| T1–T3 (no) | 12 | 0 (0%) | 12 (23.08%) | ||
| T4 (yes) | 51 | 11 (100%) | 40 (76.92%) | ||
| N stage | 2.999 | 0.083 | |||
| N0 | 45 | 5 (45.45%) | 40 (76.92%) | ||
| N+ | 18 | 6 (54.55%) | 12 (23.08%) |
Table 3
| Factor | N | High TRIM29 expression (n=44) | Low TRIM29 expression (n=36) | χ2 | P value |
|---|---|---|---|---|---|
| Gender | 12.415 | 0.000 | |||
| Male | 44 | 32 (72.73%) | 12 (33.33%) | ||
| Female | 36 | 12 (27.27%) | 24 (66.67%) | ||
| Age (years) | 24.444 | 0.000 | |||
| <60 | 16 | 0 (0%) | 16 (44.44%) | ||
| ≥60 | 64 | 44 (100%) | 20 (55.56%) | ||
| Intestinal obstruction | 6.599 | 0.010 | |||
| No | 32 | 12 (27.27%) | 20 (55.56%) | ||
| Yes | 48 | 32 (72.73%) | 16 (44.44%) | ||
| TNM Stage | 19.394 | 0.000 | |||
| I–II | 32 | 8 (18.18%) | 24 (66.67%) | ||
| III–IV | 48 | 36 (81.82%) | 12 (33.33%) | ||
| T stage (Serosa involved) | 2.566 | 0.109 | |||
| T1–T3 (no) | 28 | 12 (27.27%) | 16 (44.44%) | ||
| T4 (yes) | 52 | 32 (72.73%) | 20 (55.56%) | ||
| N stage | 19.394 | 0.000 | |||
| N0 | 32 | 8 (8.18%) | 24 (66.67%) | ||
| N+ | 48 | 36 (81.82%) | 12 (33.33%) |
Cox univariate analysis of DFS in different tumor locations
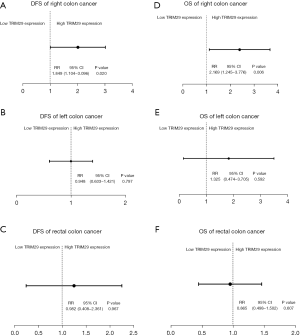
The correlation between DFS and 7 clinicopathological factors (6 clinicopathological parameters and TRIM29) was assessed by Cox regression analysis separately in patients with RCC, LCC, and RECC (Tables 4-6). In LCC patients, Cox univariate analysis revealed that gender, age, N stage, and TNM stage were prognostic factors for DFS (Table 4, P=0.044, P=0.021, P=0.000, and P=0.000, respectively). In RECC patients, none of the seven clinicopathological factors were prognostic factors for DFS (Table 5). In RCC patients, Cox univariate analysis showed that N stage, TNM stage, and TRIM29 expression levels were prognostic factors for DFS (Table 6, P=0.000, P=0.000, and P=0.020, respectively). The relative ratio (RR) of DFS for the patients with low and high TRIM29 expression is shown in Figure 2A-2C. The results revealed that, only in the RCC group, patients with high TRIM29 expression had a significantly different risk of recurrence and metastasis compared to patients with low TRIM29 expression (P=0.020).
Table 4
| Factor | Risk ratio | 95% confidence interval | P value |
|---|---|---|---|
| Gender (males were used as a reference) | |||
| Female | 0.513 | 0.268–0.982 | 0.044 |
| Age (less than 60 years was used as a reference) | |||
| ≥60 years old | 0.518 | 0.296–0.907 | 0.021 |
| Infiltration depth (T stage) (T1–3 was used as a reference) | |||
| T4 | 0.563 | 0.310–1.022 | 0.059 |
| N stage (N0 was used as a reference) | |||
| N+ | 2.968 | 1.681–5.241 | 0.000 |
| TNM stage (stage I–II was used as a reference) | |||
| Stage III | 3.392 | 1.927–5.968 | 0.000 |
| TRIM29 expression level (low TRIM29 expression was used as a reference) | |||
| High TRIM29 expression | 0.948 | 0.633–1.421 | 0.797 |
| Intestinal obstruction (no intestinal obstruction was used as a reference) | |||
| Yes | 0.892 | 0.456–1.744 | 0.739 |
Table 5
| Factor | Risk ratio | 95% confidence interval | P value |
|---|---|---|---|
| Gender (males were used as a reference) | |||
| Female | 1.044 | 0.537–2.032 | 0.898 |
| Age (less than 60 years was used as a reference) | |||
| ≥60 years old | 0.655 | 0.331–1.298 | 0.225 |
| Infiltration depth (T stage) (T1–3 was used as a reference) | |||
| T4 | 0.779 | 0.340–1.780 | 0.553 |
| N stage (N0 was used as a reference) | |||
| N+ | 1.548 | 0.758–3.162 | 0.231 |
| TNM stage (stage I–II was used as a reference) | |||
| Stage III | 1.073 | 0.469–2.456 | 0.868 |
| TRIM29 expression level (low TRIM29 expression was used as a reference) | |||
| High TRIM29 expression | 0.982 | 0.408–2.361 | 0.967 |
| Intestinal obstruction (no intestinal obstruction was used as a reference) | |||
| Yes | 0.931 | 0.386–2.241 | 0.872 |
Table 6
| Factor | Risk ratio | 95% confidence interval | P value |
|---|---|---|---|
| Gender (males were used as a reference) | |||
| Female | 0.825 | 0.498–1.367 | 0.455 |
| Age (less than 60 years was used as a reference) | |||
| ≥60 years old | 1.023 | 0.553–1.891 | 0.942 |
| Infiltration depth (T stage) (T1–3 was used as a reference) | |||
| T4 | 1.443 | 0.837–2.485 | 0.187 |
| N stage (N0 was used as a reference) | |||
| N+ | 2.903 | 1.651–5.102 | 0.000 |
| TNM stage (stage I–II was used as a reference) | |||
| Stage III | 1.704 | 1.285–2.259 | 0.000 |
| TRIM29 expression level (low TRIM29 expression was used as a reference) | |||
| High TRIM29 expression | 1.849 | 1.104–3.096 | 0.020 |
| Intestinal obstruction (no intestinal obstruction was used as a reference) | |||
| Yes | 0.647 | 0.233–1.796 | 0.403 |
Cox univariate analysis of OS in different tumor locations
The correlation between OS and the upper 7 clinicopathological factors was assessed by Cox regression analysis separately in patients with RCC, LCC, and RECC (Tables 7-9). In LCC patients, Cox univariate analysis showed that age, T stage, N stage, and TNM stage were prognostic factors for OS (Table 7, P=0.021, P=0.022, P=0.009, and P=0.008, respectively). In RECC patients, age and N stage were identified as prognostic factors for OS (Table 8, P=0.017 and P=0.020, respectively). In RCC patients, N stage, TNM stage, TRIM29 expression level, and intestinal obstruction were prognostic factors for OS (Table 9, P=0.000, P=0.000, P=0.006, and P=0.040, respectively). The results revealed that, only in the RCC group, patients with high TRIM29 expression had a significantly different risk of death compared to patients with low TRIM29 expression (Figure 2D-2F, P=0.006).
Table 7
| Factor | Risk ratio | 95% confidence interval | P value |
|---|---|---|---|
| Gender (males were used as a reference) | |||
| Female | 0.027 | 0.001–1.091 | 0.056 |
| Age (less than 60 years was used as a reference) | |||
| ≥60 years old | 0.372 | 0.160–0.863 | 0.021 |
| Infiltration depth (T stage) (T1–3 was used as a reference) | |||
| T4 | 0.373 | 0.161–0.866 | 0.022 |
| N stage (N0 was used as a reference) | |||
| N+ | 96.487 | 3.122–2,991.488 | 0.009 |
| TNM stage (stage I–II was used as a reference) | |||
| Stage III | 120.600 | 3.499–4,156.624 | 0.008 |
| TRIM29 expression level (low TRIM29 expression was used as a reference) | |||
| High TRIM29 expression | 1.325 | 0.474–3.705 | 0.592 |
| Intestinal obstruction (no intestinal obstruction was used as a reference) | |||
| Yes | 0.999 | 0.389–2.565 | 0.999 |
Table 8
| Factor | Risk ratio | 95% confidence interval | P value |
|---|---|---|---|
| Gender (males were used as a reference) | |||
| Female | 0.702 | 0.287–1.716 | 0.438 |
| Age (less than 60 years was used as a reference) | |||
| ≥60 years old | 0.222 | 0.064–0.766 | 0.017 |
| Infiltration depth (T stage) (T1–3 was used as a reference) | |||
| T4 | 0.650 | 0.216–1.955 | 0.443 |
| N stage (N0 was used as a reference) | |||
| N+ | 2.971 | 1.186–7.445 | 0.020 |
| TNM stage (stage I–II was used as a reference) | |||
| Stage III | 2.351 | 0.868–6.366 | 0.093 |
| TRIM29 expression level (low TRIM29 expression was used as a reference) | |||
| High TRIM29 expression | 0.865 | 0.498–1.502 | 0.607 |
| Intestinal obstruction (no intestinal obstruction was used as a reference) | |||
| Yes | 1.820 | 0.650–5.093 | 0.254 |
Table 9
| Factor | Risk ratio | 95% confidence interval | P value |
|---|---|---|---|
| Gender (male was used as a reference) | |||
| Female | 0.838 | 0.493–1.425 | 0.514 |
| Age (less than 60 years were used as a reference) | |||
| ≥60 years old | 1.023 | 0.538–1.948 | 0.944 |
| Infiltration depth (T stage) (T1-3 were used as a reference) | |||
| T4 | 1.598 | 0.892–2.861 | 0.115 |
| N stage (N0 were used as a reference) | |||
| N+ | 4.057 | 2.121–7.762 | 0.000 |
| TNM stage (stage I-II were used as a reference) | |||
| Stage III | 4.057 | 2.121–7.762 | 0.000 |
| TRIM29 expression level (low TRIM29 expression were used as a reference) | |||
| High TRIM29 expression | 2.169 | 1.245–3.776 | 0.006 |
| Intestinal obstruction (no intestinal obstruction was used as a reference) | |||
| Yes | 1.782 | 1.026–1.782 | 0.040 |
Kaplan-Meier analysis of the relationship between TRIM29 and DFS in patients with different tumor sites
To determine the effect of TRIM29 expression levels on prognosis in patients with different tumor sites, Kaplan-Meier survival curves and log-rank tests were performed. High TRIM29 expression was associated with poor DFS only in RCC patients, but not in LCC nor RECC patients (Figure 3A-3C). In RCC patients, the median DFS was 40 months in patients with low TRIM29 expression and 12 months in patients with high TRIM29 expression (P=0.017).
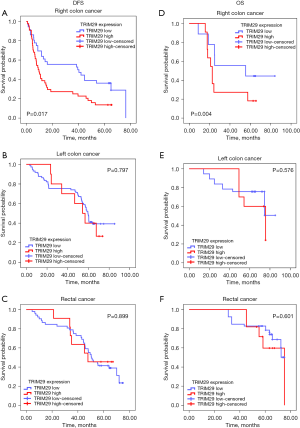
Kaplan-Meier analysis of the relationship between TRIM29 and OS in patients with different tumor sites
Kaplan-Meier survival curves and log-rank tests were conducted to investigate the relationship between OS and the levels of TRIM29 expression. The results suggested that high TRIM29 expression was associated with shorter OS only in RCC patients, but not in LCC nor RECC patients (Figure 3D-3F). In RCC patients, the median OS was 58 months in patients with low TRIM29 expression and 23 months in patients with strong TRIM29 expression (P=0.006).
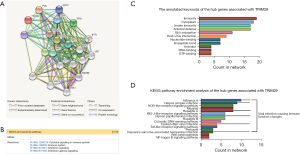
Functional annotations and the predicted signaling Reactome/KEGG pathways
A network of TRIM29 and its co-expression genes is shown in Figure 4A. The Reactome pathway analysis of the genes related with TRIM29 was performed using the String database (http://string.embl.de/) (Figure 4B). The functional enrichment pathways are cytokine signaling in immune system, immune system, interferon signaling, and interferon gamma signaling. The annotated keywords of the hub genes associated with TRIM29 is shown in Figure 4C. KEGG pathway analysis of the hub genes related to TRIM29 was performed using the String database (Figure 4D). The overlap between the Reactome/KEGG pathways enrichment and the annotated keywords analysis of the hub genes of TRIM29 suggested that TRIM29 is likely related to immune function. Considering that TRIM29 has a greater effect on RCC patients compared to either LCC or RECC patients, the correlation between TRIM29 expression and immune function was examined in colon adenocarcinoma (COAD) samples using TISIDB.
The correlation between TRIM29 expression and immune function
Analysis of the correlation between TRIM29 expression and tumor infiltrating lymphocytes using TISIDB
To clarify the correlation between TRIM29 expression and immune function, the correlation between TRIM29 expression and tumor infiltrating lymphocytes in COAD samples was examined using TISIDB (Figure 5A). Elevated TRIM29 expression was significantly associated with activated dendritic cells (Act DC), abundance of CD-56 dim cells, interdigitating dendritic cells (iDC), monocytes, neutrophils, natural killer (NK) cells, natural killer T cells (NKT), plasmacytoid dendritic cells (pDC), central memory CD4 T cells (Tcm CD4), central memory CD8 T cells (Tcm CD8), effector memory CD8 T (Tem CD8), and T-helper lymphocyte type-1 (Th1) cell infiltration in CRC (P<0.05), which was suggestive of a general increase in immune infiltration (Figure 5B-5M).
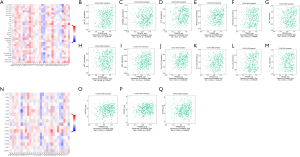
Analysis of the relationship between TRIM29 expression and chemokines using TISIDB
Chemokines play a key role in inducing immune cell infiltration, hence, the relationship between TRIM29 expression and chemokines was investigated (Figure 5N). Elevated TRIM29 was significantly associated with high expression of C-X-C motif chemokine receptor 1 (CXCR1), C-X-C motif chemokine receptor 2 (CXCR2), and C-X-C motif chemokine receptor 4 (CXCR4) (Figure 5O-5Q, P<0.05).
Analysis of the relationship between TRIM29 expression and immunostimulators using TISIDB
The correlation between TRIM29 and immunostimulators was assessed (Figure 6A). The results demonstrated that TRIM29 over-expression was significantly associated with V-Set immunoregulatory receptors (C10orf54), CD70, CD276, HERV-H LTR-associating 2 (HHLA2), and inducible T Cell costimulator ligand (ICOSLG), thereby causing immune imbalance (Figure 6B-6F, P<0.05).
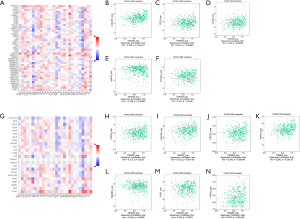
Analysis of the relationship between TRIM29 expression and immunoinhibitors using TISIDB
Immune checkpoint inhibitors are an important treatment for CRC, and thus their relationship with TRIM29 expression was explored (Figure 6G) to determine whether TRIM29 can be used as a molecular marker for the efficacy of immunotherapy. Elevated TRIM29 expression was significantly associated with CD274 (PD-L1), galectin 9 (LGALS9), programmed cell death 1 (PDCD1, PD-1), poliovirus receptor-related 2 (PVRL2), transforming growth factor beta receptor 1 (TGFBR1), T cell immunoreceptor with Ig and itim domains (TIGIT), V-set domain containing T cell activation inhibitor 1 (VTCN1), suggesting that tumors with high TRIM29 expression upregulated immune checkpoint molecules after immune stimulation to avoid immune damage (Figure 6H-6N, P<0.05).
Analysis of the correlation between TRIM29 expression levels and tumor infiltrating lymphocytes in clinical patients
H&E staining was applied to observe the tumor infiltrating lymphocytes in the above 227 formalin-fixed, paraffin-embedded CRC samples. The results revealed that high TRIM29 protein expression was correlated with high tumor-infiltrating lymphocytes (Figure 7A-7C, P=0.0053).
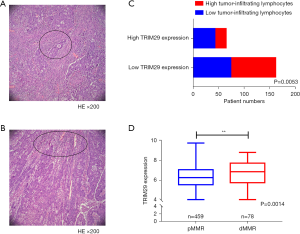
Analysis of the relationship between TRIM29 expression levels and pMMR/dMMR status using the GEO dataset
The GSE39582 dataset, consisting of 459 pMMR and 78 dMMR samples, was used to analyze the pMMR/dMMR status and TRIM29 expression level. The results revealed that dMMR patients expressed higher TRIM29 levels compared to dMMR patients (Figure 7D, P=0.0014).
Discussion
The difference in prognosis among RCC, LCC, and RECC patients is notable, with most studies confirming that RCC patients have poorer prognosis compared to patients with LCC and RECC (5,14,18-20). Furthermore, the intestinal flora, clinical manifestations, and treatment responses of patients with RCC, LCC, and RECC vary considerably (18,19). Several biomarkers, including genes, micro RNAs (miRNAs), and long non-coding RNAs (lncRNAs), have been shown to be differentially expressed and play different roles in the diagnosis and prognosis of patients with LCC and RCC. Such biomarkers include P53, excision repair cross-complementing 1 (ERCC1), forkhead box P3 (Foxp3), GNG4, and T-cell restricted intracellular antigen-1 (TIA-1) (12,21-25). However, to date, an accurate biomarker for malignancies at different tumor sites has not been identified. Our previous study was the first to demonstrated elevated expression of TRIM29 in RCC patients compared to LCC patients (14). However, this latter study did not include RECC patients. The present report is the first to analyze the prognostic value of TRIM29 in three groups of patients with different tumor locations, as well as the relationship between TRIM29 and clinicopathological features.
The current study demonstrated higher TRIM29 expression in RCC patients compared to either LCC or RECC patients. In addition, high TRIM29 expression was only associated with intestinal obstruction before surgery in LCC patients, and it was associated with higher frequency of stage TNM III–IV and younger age in RECC patients. In RCC patients, higher TRIM29 expression was associated with more clinical features (the male sex, older age, stage III–IV tumor, N+ staging, and intestinal obstruction). Furthermore, high TRIM29 expression was significantly associated with an increased risk of recurrence/metastasis and death only in RCC patients.
TRIM29 is a number of the TRIM family that plays an important regulatory role in immune functions (26,27). Indeed, TRIM29 has been reported to act as an oncogene in many cancers, such as CRC, breast cancer, and lung cancer (28-31). It has been shown to affect immune function by regulating interferon levels and ubiquitination (26,27,32). Bioinformatics analysis suggested that TRIM29 may regulate tumor immunity in CRC. Therefore, this study examined the relationship between TRIM29 expression and tumor immune function in COAD patients by using TISIDB. The results revealed that TRIM29 may be involved in a general increase in immune infiltration and overexpression of chemokines. A report has suggested that the immune microenvironment in patients with CRC on the right side is characterized by increased infiltration of immune cells and higher interferon-γ signatures (33). In agreement with previous literature (26,27), our results suggested that RCC patients are more likely to overexpress TRIM29, leading to increased immune infiltration and overexpression of chemokines. Furthermore, certain immunostimulators were overexpressed with upregulated TRIM29 expression. Interestingly, certain immunoinhibitors were also upregulated with TRIM29 overexpression. It is plausible that tumors with high TRIM29 expression upregulate immune checkpoint molecules after immune stimulation to avoid immune damage.
The above data verified that TRIM29 plays a significant role in RCC patients, resulting in a general increase in immune infiltration that may lead to immune imbalance. TRIM29 may act as a novel biomarker and a potential therapeutic target in RCC patients. However, larger prospective studies are warranted to confirm these results.
Acknowledgments
Funding: This work was supported by the Youth Foundation of Hebei Province (No. H2020206394).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-365/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-365/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-365/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The collection of all human samples in this study was approved by ethics board of Hebei Medical University Fourth Affiliated Hospital (No. 2017MEc089). Informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med 1990;113:779-88. [Crossref] [PubMed]
- Tamas K, Walenkamp AM, de Vries EG, et al. Rectal and colon cancer: Not just a different anatomic site. Cancer Treat Rev 2015;41:671-9. [Crossref] [PubMed]
- Xu JM. Difference of colon cancer and rectal cancer-from the view of an oncological physician. Zhonghua Zhong Liu Za Zhi 2010;32:321-3. [PubMed]
- Price TJ, Beeke C, Ullah S, et al. Does the primary site of colorectal cancer impact outcomes for patients with metastatic disease? Cancer 2015;121:830-5. [Crossref] [PubMed]
- Aggarwal H, Sheffield KM, Li L, et al. Primary tumor location and survival in colorectal cancer: A retrospective cohort study. World J Gastrointest Oncol 2020;12:405-23. [Crossref] [PubMed]
- Shida D, Inoue M, Tanabe T, et al. Prognostic impact of primary tumor location in Stage III colorectal cancer-right-sided colon versus left-sided colon versus rectum: a nationwide multicenter retrospective study. J Gastroenterol 2020;55:958-68. [Crossref] [PubMed]
- Missiaglia E, Jacobs B, D'Ario G, et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol 2014;25:1995-2001. [Crossref] [PubMed]
- Papagiorgis PC, Zizi AE, Tseleni S, et al. The pattern of epidermal growth factor receptor variation with disease progression and aggressiveness in colorectal cancer depends on tumor location. Oncol Lett 2012;3:1129-35. [Crossref] [PubMed]
- Schell MJ, Yang M, Teer JK, et al. A multigene mutation classification of 468 colorectal cancers reveals a prognostic role for APC. Nat Commun 2016;7:11743. [Crossref] [PubMed]
- Chen KH, Shao YY, Chen HM, et al. Primary tumor site is a useful predictor of cetuximab efficacy in the third-line or salvage treatment of KRAS wild-type (exon 2 non-mutant) metastatic colorectal cancer: a nationwide cohort study. BMC Cancer 2016;16:327. [Crossref] [PubMed]
- Song J, Yang J, Lin R, et al. Molecular heterogeneity of guanine nucleotide binding-protein gamma subunit 4 in left- and right-sided colon cancer. Oncol Lett 2020;20:334. [Crossref] [PubMed]
- Dong W, Li N, Pei X, et al. Differential expression of DUSP2 in left- and right-sided colon cancer is associated with poor prognosis in colorectal cancer. Oncol Lett 2018;15:4207-14. [Crossref] [PubMed]
- Han J, Zhao Z, Zhang N, et al. Transcriptional dysregulation of TRIM29 promotes colorectal cancer carcinogenesis via pyruvate kinase-mediated glucose metabolism. Aging (Albany NY) 2021;13:5034-54. [Crossref] [PubMed]
- Han J, Zhang X, Yang Y, et al. Screening and Identification of Differentially Expressed Genes Expressed among Left and Right Colon Adenocarcinoma. Biomed Res Int 2020;2020:8465068. [Crossref] [PubMed]
- Krzystek-Korpacka M, Zawadzki M, Kapturkiewicz B, et al. Subsite heterogeneity in the profiles of circulating cytokines in colorectal cancer. Cytokine 2018;110:435-41. [Crossref] [PubMed]
- Takasu C, Nishi M, Yoshikawa K, et al. Impact of sidedness of colorectal cancer on tumor immunity. PLoS One 2020;15:e0240408. [Crossref] [PubMed]
- Christodoulidis G, Spyridakis M, Symeonidis D, et al. Clinicopathological differences between right- and left-sided colonic tumors and impact upon survival. Tech Coloproctol 2010;14:S45-7. [Crossref] [PubMed]
- Vaish V, Kim J, Shim M. Lentivirus-mediated somatic recombination and development of a novel mouse model for sporadic colorectal cancer. Genes Chromosomes Cancer 2016;55:577-90. [Crossref] [PubMed]
- Mangone L, Pinto C, Mancuso P, et al. Colon cancer survival differs from right side to left side and lymph node harvest number matter. BMC Public Health 2021;21:906. [Crossref] [PubMed]
- Mik M, Berut M, Dziki L, et al. Right- and left-sided colon cancer - clinical and pathological differences of the disease entity in one organ. Arch Med Sci 2017;13:157-62. [Crossref] [PubMed]
- Li KZ, Yin YX, Tang YP, et al. Construction of a long noncoding RNA-based competing endogenous RNA network and prognostic signatures of left- and right-side colon cancer. Cancer Cell Int 2021;21:211. [Crossref] [PubMed]
- De Renzi G, Gaballo G, Gazzaniga P, et al. Molecular Biomarkers according to Primary Tumor Location in Colorectal Cancer: Current Standard and New Insights. Oncology 2021;99:135-43. [Crossref] [PubMed]
- Kanno H, Miyoshi H, Yoshida N, et al. Differences in the immunosurveillance pattern associated with DNA mismatch repair status between right-sided and left-sided colorectal cancer. Cancer Sci 2020;111:3032-44. [Crossref] [PubMed]
- Hirabayashi S, Hayashi M, Nakayama G, et al. The Significance of Molecular Biomarkers on Clinical Survival Outcome Differs Depending on Colon Cancer Sidedness. Anticancer Res 2020;40:201-11. [Crossref] [PubMed]
- Xing J, Zhang A, Zhang H, et al. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat Commun 2017;8:945. [Crossref] [PubMed]
- Li Q, Lin L, Tong Y, et al. TRIM29 negatively controls antiviral immune response through targeting STING for degradation. Cell Discov 2018;4:13. [Crossref] [PubMed]
- Sun J, Zhang T, Cheng M, et al. TRIM29 facilitates the epithelial-to-mesenchymal transition and the progression of colorectal cancer via the activation of the Wnt/beta-catenin signaling pathway. J Exp Clin Cancer Res 2019;38:104. [Crossref] [PubMed]
- Xu W, Chen B, Ke D, et al. TRIM29 mediates lung squamous cell carcinoma cell metastasis by regulating autophagic degradation of E-cadherin. Aging (Albany NY) 2020;12:13488-501. [Crossref] [PubMed]
- Li W, Xue H, Li Y, et al. ATDC promotes the growth and invasion of hepatocellular carcinoma cells by modulating GSK-3beta/Wnt/beta-catenin signalling. Clin Exp Pharmacol Physiol 2019;46:845-53. [Crossref] [PubMed]
- Qiao HY, Zhang Q, Wang JM, et al. TRIM29 regulates the SETBP1/SET/PP2A axis via transcription factor VEZF1 to promote progression of ovarian cancer. Cancer Lett 2022;529:85-99. [Crossref] [PubMed]
- Dou Y, Xing J, Kong G, et al. Identification of the E3 Ligase TRIM29 as a Critical Checkpoint Regulator of NK Cell Functions. J Immunol 2019;203:873-80. [Crossref] [PubMed]
- Zhang L, Zhao Y, Dai Y, et al. Immune Landscape of Colorectal Cancer Tumor Microenvironment from Different Primary Tumor Location. Front Immunol 2018;9:1578. [Crossref] [PubMed]


