
Exploring potential targets of Actinidia chinensis Planch root against hepatocellular carcinoma based on network pharmacology and molecular docking and development and verification of immune-associated prognosis features for hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) has the characteristics of insidious onset, strong invasiveness, and easy metastasis and recurrence. For these reasons, it has one of the highest incidence and death rates of all malignant tumors worldwide. Therefore, liver cancer patients tend to have a poor prognosis. At present, the main treatment means for liver cancer include surgery, targeted therapy, and immunotherapy, among others. However, in practical clinical applications, due to factors such as resistance to targeted drugs and immune-related toxicity, patients have limited treatment efficacy, which also affects their quality of life (1,2). Therefore, it is imperative to explore complementary and alternative medicines for the treatment of HCC.
In recent years, traditional Chinese medicine has been used as an increasingly vital component of the comprehensive treatment of HCC. It has unique advantages in improving the symptoms of patients, enhancing immune function, and prolonging survival (3,4). In a literature review on the anti-cancer effects of traditional Chinese medicine, we found that the root of Actinidia chinensis Planch (ACP) has a wide range of anti-cancer activities, especially in digestive tract tumors (5-9). Therefore, the root of ACP is expected to bring new hope for conquering cancer in clinical practice. The ACP root, also known as Radix Actinidia chinensis, belongs to the Actinidiaceae family and has a long history. It has been recorded from Erya to Compendium of Materia Medica more than 2,000 years ago in China. It mainly grows in the southern region of China. Its taste is salty, sour, astringent, cool in nature, and non-toxic. It has the functions of tonifying the spleen and eliminating dampness, dissolving phlegm and dissipating stagnation, heat-clearing and detoxifying, improving blood circulation and reducing swelling, and heat dissipation to stop bleeding. The root of ACP contains a variety of medicinal ingredients, such as triterpenoids, flavonoids, enquinones, and alkaloids, among others (10). Currently, there are many compound preparations containing ACP root used in the folk treatment of cancer, such as Jiedu Sangen Decoction, which is composed of roots of Polygonum cuspidatum Sieb. et Zucc., Root of Adina rubella Hance, and Radix Actinidia chinensis (11). The root of ACP has good curative effect in inhibiting the proliferation, migration, invasion and promoting apoptosis of liver cancer cells. For example, Fang et al. applied Actinidia chinensis Planch root extract (acRoots) (10 mg/mL) to normal liver epithelial cell line (L02) and human HCC cell lines (Hep3B, HepG2 and SMMC7721). It was found that acRoots was not cytotoxic to L02 cells, but it significantly inhibited the proliferation of HCC cells (12). The scholar also found that acRoots can inhibit the proliferation and metastasis of liver cancer cells by down-regulating EP3 expression and blocking the DLX2/TARBP2/JNK/AKT signaling pathway (13,14). Hou et al. found that the extract from root of Actinidia chinensis (ERAC) could effectively attenuate the cell growth of human hepatoma cell lines LM3, HepG2, 97H, 97L, SMMC-7721, Hep3B, Huh7 and HCCC-9810, which may be due to ERAC inhibiting the proliferation of highly metastatic hepatoma cells through the gene encoding laminin subunit beta-3 (LAMB3) (15). It can be seen that the root of ACP has good curative effect and certain safety in anti-hepatocellular carcinoma. However, due to the synergistic effect of various chemical components and multiple targets in ACP root, its curative effect is more diversified, and it is as a result of these characteristics that it has a complex mechanism, which has not yet been fully elucidated. This limits the research, development, and clinical application of ACP root. For this reason, it is necessary to further study the underlying molecular mechanisms of its potential anti-HCC effects.
Many scholars believe that the main pathogenesis of HCC is asthenia in origin and asthenia in superficiality, of which “spleen deficiency” is the root (16). The spleen has an important immune function in our body. “Spleen deficiency” is closely related to the decline of the body’s immune function, which is also consistent with modern medicine. Patients with liver cancer usually have low immune function, which weakens the immune system’s ability to fight cancer cells. This results in immune escape, leading to tumor progression, invasion, or metastasis. It is well known that HCC immune escape is an important feature of HCC (17). Currently, in clinical practice, targeting programmed cell death ligand 1 (PD-L1), programmed cell death 1 (PD-1) and cytotoxic Tlymphocyte antigen-4 (CTLA-4) targets, inhibiting their expression to enhance the body’s immune response and/or destroy tumors immunosuppression and other pathways to delay the progression of tumors (18). In addition, PD-L1/PD-1 and CTLA-4 play important roles in predicting the survival time of HCC patients and evaluating the efficacy of immunotherapy, but their sensitivity and specificity in HCC cells are not high. Therefore, it is very necessary to discover new immune-related tumor markers and new therapeutic targets in liver cancer.
According to the “treating deficiency syndrome with tonifying methods” in traditional Chinese medicine, “spleen deficiency” requires replenishing the spleen (19). Therefore, improving the immune function of HCC patients is a key part of effectively preventing the malignant growth of liver cancer cells. Numerous studies have shown that traditional Chinese medicine methods, such as invigorating the spleen, supplementing qi, nourishing qi, and solidifying, can promote the development of immune cells and immune organs and block immune checkpoints, among other processes, to improve the body’s ability to respond to tumors (20-22). It has been demonstrated that the ACP root, which has the effect of strengthening the spleen, plays a critical role in regulating the immune system of tumor patients. To sum up, in light of the traditional Chinese medicine theory, we studied the anti-HCC mechanism of ACP root from the perspective of immune regulation, which will open up new research ideas and therapeutic approaches for ACP root in the prevention and treatment of HCC.
Therefore, in this study, we screened the active components of ACP and related targets for the treatment of liver cancer by network pharmacology, and clarified the relationship between ACP and HCC by constructing a network of “active components of drugs-targets-liver cancer“. We then used bioinformatics technology to screen out genes with significantly different expression in liver cancer compared with the normal group, and analyzed target enrichment functions and pathways. After that, we constructed a prognosis model of immune-related genes, obtained the key targets of ACP in the treatment of liver cancer by analyzing the prognostic value of liver cancer, and molecularly docked them with the corresponding components. The goal of this study is to explore the effective components and potential targets of ACP in the treatment of liver cancer, and to find immune-related biomarkers that can be used to predict the prognosis of HCC. The clinical significance of this study provides a theoretical basis and a new opportunity for the development and application of ACP in the treatment of liver cancer, and it also lays the foundation for useful indicators to predict the prognosis of HCC patients and candidates for HCC treatment. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-398/rc).
Methods
Data collection
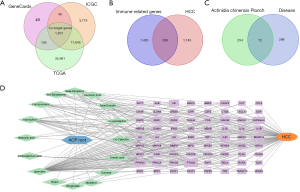
We use the Traditional Chinese Medicine Systems Pharmacology database (TCMSP, https://tcmsp-e.com/, updated 2022-03-14), SymMAP database (http://www.symmap.org/), and HERB database (http://herb.ac.cn/) to obtain the active components and target genes of Actinidia chinensis Planch root. We searched for “Hepatocellular carcinoma” in the GeneCards database (https://www.genecards.org/, updated 2022-03-14), and screened out those with a correlation score greater than 5, a total of 1,757 liver cancer genes were obtained, and then download liver cancer-related genes from the TCGA database (https://portal.gdc.cancer.gov/, updated 2022-03-14) and the ICGC database (https://dcc.icgc.org/, updated 2022-03-14), and the common liver cancer genes in these three databases were obtained. We derived immune-related genes from the “Gene Lists” module in the ImmPort database (https://www.immport.org/home). Based on ACP target genes, liver cancer common genes and immune-related genes, the network of “ACP-target-HCC” was subsequently constructed. At the same time, we also downloaded transcriptome expression data and clinical information from the latter two databases (TCGA database: 374 HCC cases and 50 normal cases, ICGC database: 243 HCC cases and 202 normal cases), which were used in the construction and validation of liver cancer prognosis models. Tumor immune cell infiltration data was obtained from the TIMER2.0 website (http://timer.cistrome.org/), and its data were applied to analyze potential links between prognostic models and immune cell infiltration. Finally, we obtained the protein structures of the core target genes from the PDB database (https://www.rcsb.org/) and the SDF format files of the small molecule ligands from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), which were then utilized for molecular docking. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Determination of the core targets and network construction
We entered “Actinidia chinensis Planch” in the TCMSP database to screen the active ingredients of ACP with oral bioavailability (OB) ≥30% and drug-likeness (DL) ≥0.18, and the corresponding target genes were obtained. We still considered active ingredients that did not meet this requirement but still had significant biological activity as well as those reported in the literature. Molsoft website (http://molsoft.com/mprop/) and preADMET website (https://preadmet.bmdrc.kr/) were used to evaluate the DL value of the active ingredient, and then the target genes of the active components of ACP were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/).
The common genes of liver cancer were extracted from the GeneCards, TCGA, and ICGC databases, and the immune-related target genes of ACP root were extracted from the common genes. The final screening results were illustrated by a Venn diagram drawn by the “VennDiagram” R package using R software (version 4.1.3, https://cran.r-project.org/src/base/R-4/R-4.1.3.tar.gz). We employed Cytoscape 3.9.0 software to construct an “active component-target-disease” network for the target genes that ACP active components act on in immune-related liver cancer. We applied the “limma” R package to perform differential analysis on the transcriptome data of the liver cancer group and the normal group in the TCGA database, in order to obtain differentially expressed genes, where P<0.05 and |logFC|>2 were fixed as filter conditions. Then, we used the “pheatmap” and “ggplot2” R packages to draw a heat map and a grouped boxplot, respectively, for the differential genes. We set up a protein-protein interaction (PPI) network for obviously different genes via the “multiple proteins” module in the STRING database (https://string-db.org/). The screening conditions were set as human species, minimum required interaction score of 0.4, and the item “hide disconnected nodes in the network” was selected. Finally, we exported the PNG image and saved it as a TVS file, and the immune-related core target genes that ACP acts on in HCC were obtained.
Functional analysis of core targets
In order to analyze the status of active compounds in ACP roots and the function of core immune-related target proteins, we performed Gene Ontology (GO) enrichment analysis on the intersection genes to gain a preliminary understanding of their biological functions, pathways, and cellular localization. We utilized the “colorspace”, “stringi”, “DOSE”, “clusterProfiler”, and “pathview” R packages to draw the top 10 regulations in the 3 biological processes, the filter condition was set to P<0.05, and bubble charts and bar charts were drawn. In order to understand the pathways which are significantly altered during ACP root treatment of liver cancer, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. P<0.05 was the screening criterion, and “enrichplot”, “ggplot2”, and “pathview” packages were used to draw the top 30 KEGG pathways, which were displayed in bubble charts and bar charts.
Establishment and validation of predictive risk models
To determine the association between core target genes in ACP treatment for HCC and the poor prognosis of patients, based on the TCGA and ICGC databases, we extracted the overall survival (OS) time, survival status, and mRNA expression of target genes from eligible cases that had complete basic clinical information and a survival time that was not 0 days. The “limma” R package was used to merge the gene expression data and the corresponding survival data. The “survival” package was adopted for univariate Cox regression analysis. Subsequently, we selected genes with P<0.05 for LASSO analysis. Finally, the λ with the smallest partial likelihood deviation was selected as the optimal λ, and the predicted genes were utilized to build the prognostic model. The risk score formula was , where Coefi is the risk coefficient and the amount of expression of each gene is represented by xi. We used the “survival” and “survminer” R packages to carry out Kaplan-Meier survival analysis on high- and low-risk groups, which were divided according to the median value of risk, and plotted the corresponding survival curve graphs. Meanwhile, to further evaluate the performance of the prognostic model, we used the “survivalROC” package to perform receiver operating characteristic (ROC) curve analysis. Subsequently, we utilized the Liver Cancer-NCC-JP dataset in the IGCC database to verify the predictive capability of the model we constructed. In addition, we combined clinically relevant information, including age, gender, and pathological stage, among others, to be included in univariate Cox and multivariate Cox analyses on the training set (TCGA database) as well as the test set (ICGC database) of the prognostic model in progress, with the purpose of investigating whether this model can determine the independent predictive factors for OS. We also drew a clinical correlation heat map to intuitively clarify the relationship between the prognostic pattern’s risk score and the characteristics of liver cancer patients, such as age, gender, and grade.
Correlation analysis of the prediction model with immune cells and immune checkpoint genes
We assessed the correlation between our prognostic model’s risk grade and the contents of 6 immune-infiltrating cells, including B cells, CD4+ T cells, macrophages, CD8+ T cells, neutrophils, and myeloid dendritic cells, with the aim of understanding the relationship between this model and tumor immune cells in HCC patients. The final results were displayed in a correlation scatterplot, where Pearson correlation coefficient (Cor) >0 indicated a positive correlation, otherwise Cor <0 signified a negative correlation, and a P<0.05 was considered to be significant. We used the “ggplot2” R package to determine the reciprocity of the risk score of the prognostic model and the expression levels of common immune checkpoint genes, including CTLA-4, transforming growth factor beta 1 (TGFB1), transforming growth factor beta receptor 1 (TGFBR1), PD-1, interleukin 10 (IL-10), lymphocyte-activation gene 3 (LAG3), cluster of differentiation 96 (CD96), indoleamine 2,3-dioxygenase 1 (IDO1), and PD-L1. Finally, we performed gene set enrichment analysis (GSEA) to explore latent signaling paths associated with the genes in the prognostic model by utilizing GSEA_4.2.3 software (http://www.gsea-msigdb.org/gsea/downloads.jsp/GSEA_Win_4.2.3-installer.exe).
Molecular docking verification
We performed molecular docking in order to further verify the close connection between active compounds in ACP root and prognostic model genes. We downloaded the SDF format files of the 2D structures of small molecule ligands from the PubChem database, imported them into the ChemBio3D Ultra 14.0 software to convert them into a 3D structure, and saved them as a mol2 format file. We then used AutoDockTools software to calculate the atom type and charge of the small molecule ligand, and saved it as a pdbqt format file. We obtained the 3D protein structures of the prognostic model genes from the PDB database (https://www.rcsb.org/), downloaded them in PDB format, imported them into PyMOL software to dehydrate the target original protein conformation and remove small molecule ligands, and saved them as pdb format file. Then, after hydrogenation and other processing were carried out with AutoDockTools, the files were converted into pdbqt format, the center of the receptor structure was selected to draw a box, and the docking parameters such as conformation search method were set for molecular docking calculation. We selected important medicinal components and key targets for molecular docking verification. Finally, we applied Vina software for molecular docking, and visualized the docking outcomes using PyMOL software.
Statistical analysis
In this study, the “limma” R package was applied to analyze the gene expression differences between the liver cancer group and the normal group. We used the LASSO regression algorithm to construct the risk prognostic model, and then employed the Kaplan-Meier method to draw the survival curve and use the Log-rank test for survival analysis. We calculated the area under the curve (AUC) of the ROC curve by the “survivalROC” package. Afterwards, univariate and multivariate Cox analyses were employed to assess the feasibility of risk score as an independent predictor of OS. We utilized the Pearson correlation test to analyze the correlation between the prognostic model and the six types of immune cells, and the Spearman correlation test for the relationship between the prognostic model and the expression of immune checkpoints. In the above analysis, all statistical analyses were performed using R software version 4.1.3, and P<0.05 was considered to be statistically significant.
Results
Screening results of pharmacodynamic components and potential targets of ACP root
We finally screened 16 active chemical components (see Table 1), most of which were triterpenoids and flavonoids, and predicted 326 potential targets. We collected a total of 1,501 HCC genes from 3 databases (Figure 1A) and acquired 2,483 immune-related genes, of which 358 genes were related to liver cancer and immunity (Figure 1B). The Venn diagram showed a total of 72 immune-related target genes that the active components of ACP root act on in liver cancer (Figure 1C). We used Cytoscape software to form a network diagram of “active ingredients-target-liver cancer” composed of 16 active compounds in ACP root and 72 target genes (Figure 1D). The main effective components of ACP in the treatment of HCC include quercetin, (+)-catechin, ent-epicatechin, kaempferol, (−)-epicatechin, beta-sitosterol, and formononetin, among others.
Table 1
| Mol ID | Molecule name | OB (%) | DL |
|---|---|---|---|
| MOL000073 | Ent-epicatechin | 48.96 | 0.24 |
| MOL000098 | Quercetin | 46.43 | 0.28 |
| MOL000105 | Protocatechuic acid | 25.37 | 0.04 |
| MOL000263 | Oleanolic acid | 29.02 | 0.76 |
| MOL000357 | Sitogluside | 20.63 | 0.62 |
| MOL000358 | Beta-sitosterol | 36.91 | 0.75 |
| MOL000359 | Sitosterol | 36.91 | 0.75 |
| MOL000392 | Formononetin | 69.67 | 0.21 |
| MOL000422 | Kaempferol | 41.88 | 0.24 |
| MOL000471 | Aloe-emodin | 83.38 | 0.24 |
| MOL000492 | (+)-catechin | 54.83 | 0.24 |
| MOL000511 | Ursolic acid | 16.77 | 0.75 |
| MOL000842 | Sucrose | 7.17 | 0.23 |
| MOL001691 | Ascorbic acid | 13.34 | 0.04 |
| MOL002268 | Rhein | 47.07 | 0.28 |
| MOL006820 | (−)-epicatechin | 28.93 | 0.24 |
OB, oral bioavailability; DL, drug-likeness.
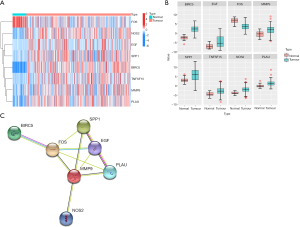
After differential classification with TCGA database, we obtained 8 differentially expressed genes. From the heat map and grouped boxplot, we observed that 7 genes were up-regulated in liver cancer tissues, namely epidermal growth factor (EGF), matrix metallopeptidase 9 (MMP9), baculoviral inhibitor of apoptosis repeat-containing protein 5 (BIRC5), nitric oxide synthase (NOS2), plasminogen activator urokinase (PLAU), Tumor necrosis factor superfamily member 15 (TNFSF15), and secreted phosphoprotein 1 (SPP1), while 1 gene was down-regulated in HCC tissues, namely the c-fos protein (FOS) (Figure 2A,2B). We used the STRING database to build a PPI network, which included 8 nodes, 12 interaction relationships, and an average node degree of 3 (Figure 2C). Finally, 7 genes (EGF, MMP9, BIRC5, NOS2, PLAU, FOS, SPP1) were obtained.
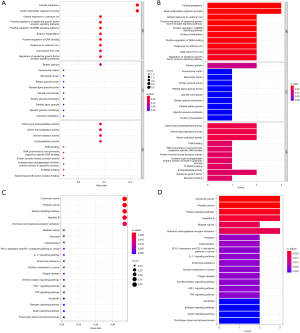
A total of 365 related items were obtained by GO enrichment, including 333 biological process (BP) items, which were mainly related to female pregnancy, multi-multicellular organism process, cellular response to cadmium ion, positive regulation of ERBB signaling pathway, and positive regulation of EGF receptor signaling pathway, among others. Only 1 cellular component (CC) entry was found, namely tertiary granule. At the same time, we obtained 30 molecular function (MF) items, mainly including serine-type endopeptidase activity, serine-type peptidase activity, serine hydrolase activity, RNA polymerase II core promoter sequence-specific DNA binding, protein tyrosine kinase activator activity, and FMN binding, among others. According to P<0.05, the top 10 items of each of the 3 branches were selected to make bubble charts and bar graphs, as shown in Figure 3A and Figure 3B. Through KEGG pathway enrichment analysis, according to P<0.05, we obtained a total of 26 pathways, mainly including colorectal cancer, prostate cancer, relaxin signaling pathway, hepatitis B, bladder cancer, chemical carcinogenesis-receptor activation, pertussis, leishmaniasis, PD-L1 expression, and PD-1 checkpoint pathway in cancer, among others. Then, we made bubble charts and histograms of the relevant signaling pathways, as shown in Figure 3C and 3D.
The results of prognostic analysis
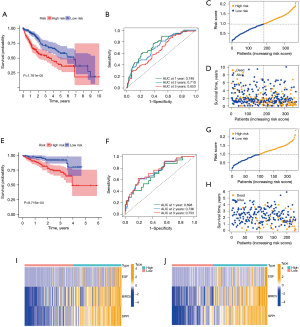
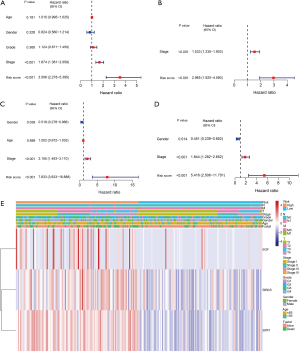
In order to further analyze and screen out the main target genes of the root of ACP, we used the liver cancer data in TCGA database as the training set, and performed univariate Cox regression analysis on the training set to find 5 survival-related genes (Figure 4A; P<0.05). After LASSO regression analysis, 3 genes were identified to construct a prognostic model (Figure 4B,4C), including EGF, BIRC5, and SPP1 (P<0.05). These 3 genes were all high-risk genes and were significantly up-regulated in liver cancer tissues. The risk score was calculated as follows: risk score = (0.160956867246387 × gene EGF expression value) + (0.188393314587069 × gene BIRC5 expression value) + (0.0901620889436574 × gene SPP1 expression value). In the training set, the Kaplan-Meier survival analysis curves revealed that the survival rate of the high-risk group was lower than that of the low-risk group (P=1.761e-05; Figure 5A), indicating that the model has a certain latent capacity in forecasting the prognosis of HCC sufferers. Besides, the area under the curve (AUC) values for OS at 1 year, 2 years, and 3 years were, respectively, 0.749, 0.710, and 0.653 in the training set (Figure 5B). Simultaneously, the corresponding risk score distribution curve and survival status diagram also displayed differences (Figure 5C,5D). The model was also validated to have certain predictive value in the training set (P=8.716e-04; Figure 5E), where the AUC values for OS at 1, 2, and 3 years were 0.698, 0.736, and 0.753 in the test set (Figure 5F). Each AUC value was larger than 0.6, clearly indicating that this model has good potential in calculating the prognosis of HCC patients. And the risk score distribution curve and survival status diagram of the test set, as shown in Figure 5G and Figure 5H. In addition, we also plotted a heat map of the clinical correlations of the training and test sets, as shown in Figure 5I and Figure 5J. We further utilized univariate and multivariate Cox regression analysis to estimate the independent predictive capacity of the prognosis of immune-related genes of ACP targets. The final results revealed that stage and risk grades were evidently correlated with the OS of patients in both the training and test sets (both P<0.001). In addition, gender showed a prominent association with patient OS in both the univariate and multivariate analyses in the test set (P<0.05; Figure 6A-6D). At the same time, through the clinical correlation heat map, we visually observed that the prognostic model’s risk score, to some extent, had a correlation with the age, gender, pathological grade, tumor TMN stage, clinical stage, and other characteristics of liver cancer patients (Figure 6E). Therefore, we believe that this prognostic model can serve as an independent predictive marker.

The results of immunomodulatory analysis
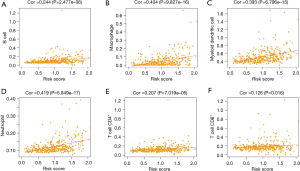
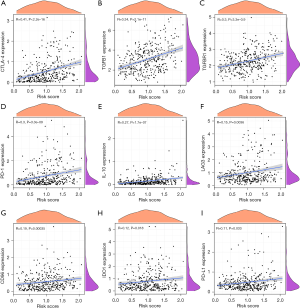
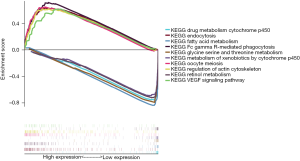
We further explored the potential association between the immune-related prognostic model of ACP targets and immune cell infiltration, as shown in Figure 7A-7F. The results were as follows: B cells (Cor =0.244, P=2.477e-06), macrophages (Cor =0.404, P=9.827e-16), myeloid dendritic cells (Cor =0.393, P=6.796e-15), neutrophils (Cor =0.419, P=5.849e-17), CD4+ T cells (Cor =0.207, P=7.019e-05), CD8+ T cells (Cor =0.126, P=0.016). These 6 immune cells all possessed a notable positive correlation with the risk score of the ACP target immune-related prognostic model, and with higher risk scores, the levels of these infiltrating immune cells also increased. In addition, we analyzed the potential relationship between the risk scores of models representing prognosis and the expression levels of familiar immune-checkpoint genes, as shown in Figure 8A-8I. The results were as follows: CTLA-4 (R =0.41, P<2.2e-16 ), TGFB1 (R =0.34, P=2.1e-11), TGFBR1 (R =0.3, P=3.3e-09), PD-1 (R =0.3, P=3.3e-09), IL-10 (R =0.27, P=1.7e-07), LAG3 (R =0.15, P=0.0036), CD96 (R =0.19, P=0.00035), IDO1 (R =0.12, P=0.018), and PD-L1 (R =0.11, P=0.033). The results showed that the expression levels of these immunosuppressive genes gradually increased as the risk score increased, and the 2 were positively correlated (P<0.05). At the same time, the outcomes of GSEA indicated that the prognostic model ranked the top 5 pathways in the high-risk group, including Fc gamma R-mediated phagocytosis, oocyte meiosis, endocytosis, regulation of actin cytoskeleton, and VEGF signaling pathway, while in the low-risk group, the top 5 pathways included drug metabolism cytochrome P450, retinol metabolism, fatty acid metabolism, metabolism of xenobiotics by cytochrome P450, and glycine, serine, and threonine metabolism (Figure 9).
Molecular docking verification
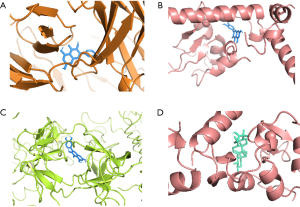
We applied AutoDockTools software to conduct molecular docking of the 3 target genes EGF, BIRC5, and SPP1 with the active components of ACP roots that act on them (Table 2). It is generally recognized that the lowest binding energy is less than or equal to −5.0 kcal/mol, indicating that the pharmacodynamic molecule has a better capability to combine with the protein, and the lower the binding energy, the better the binding ability between the molecule and the protein. In our study, the results of molecular docking exhibited that their binding energies were all ≤−5.0 kcal/mol, which illustrated that they all had an excellent binding ability. The detailed results are as follows: quercetin-SPP1 −8.2 kcal/mol, quercetin-BIRC5 −8.1 kcal/mol, quercetin-EGF −7.9 kcal/mol, ursolic acid (UA)-BIRC5 −6.9 kcal/mol, and quercetin-SPP1 had the smallest binding energy. We used PyMOL software to visualize the molecular docking results of quercetin-SPP1, quercetin-BIRC5, quercetin-EGF, and UA-BIRC5 (Figure 10A-10D).
Table 2
| Molecular docking mode | Binding energy (kcal/mol) |
|---|---|
| Quercetin-SPP1 | −8.2 |
| Quercetin-BIRC5 | −8.1 |
| Quercetin-EGF | −7.9 |
| Ursolic acid-BIRC5 | −6.9 |
The darker the color in Table 2, the stronger the binding ability of this docking model. ACP, Actinidia chinensis Planch; SPP1, secreted phosphoprotein 1; BIRC5, baculoviral inhibitor of apoptosis repeat-containing protein 5; EGF, epidermal growth factor.
Discussion
It is well known that the immune system of the human body can monitor, identify, and eliminate the vast majority of early cancer cells. Therefore, enhancing the immune function of the body can hinder the occurrence, development, metastasis, and recurrence of cancer (23). HCC is often described as “liver accumulation” or “accumulation” in ancient Chinese medical books. In traditional Chinese medicine theory, liver cancer is mainly caused by deficiency of righteous qi, especially “spleen deficiency”, which is described in the book Jingyue Quanshu: “People who are deficient in the spleen and kidney, and those who are weak and disordered, tend to have accumulated diseases” (24). “Spleen deficiency” in traditional Chinese medicine is closely related to abnormal immune function in the internal environment of liver cancer. ACP root, as a representative traditional Chinese herbal medicine for invigorating spleen and qi, has many years of history in the treatment of liver cancer. However, the underlying mechanism between ACP root and HCC is still not completely elucidated. In our study, we speculated that the root of ACP affects liver cancer through immunomodulatory effects.
We used network pharmacology to establish a network of “drug active ingredients-target genes-HCC”, then obtained the transcriptome data and clinical information from TCGA and ICGC databases, and finally derived the 3 genes EGF, BIRC5, and SPP1. They are not only the hub genes of ACP root against HCC, but are also associated with the prognosis of liver cancer. Relevant research has discovered that EGF is a cancer-promoting factor in a variety of malignant tumors. It is highly expressed in liver cancer and can take part in the occurrence and development of liver cancer through a variety of pathways, such as EGF/EGFR, MAPK, and PI3K bypass signal transduction, as well as through participation in drug resistance, promotion of angiogenesis, and other mechanisms to promote the metastasis and malignant proliferation of liver cancer (25-29). In our study, it was found that quercetin, which is one of the active ingredients in the root of ACP, can act on EGF targets. Quercetin is a polyphenolic flavonoid, also known as 3,30,40,5,7-pentahydroxyflavone. Research on this compound in the field of HCC is relatively mature, with studies showing that it can play a role by blocking the JAK2/STAT3 signaling pathway, inhibiting the Akt-mTOR pathway, hindering the activation of NF-κB, and impeding PI3K/p53/COX-2 (30,31). In addition, quercetin down-regulates the expression of EGF to restrain the proliferation of cancer cells, which has also been demonstrated in previous literature (32,33). Nevertheless, this mechanism has not been reported in liver cancer, and further experiments are needed for verification.
BIRC5, also known as survivin, is one of the key genes in the inhibitor of apoptosis protein (IAP) family (34). A great deal of studies have shown that BIRC5, which is widely and highly expressed in various malignant tumor cells, is an accomplice to help tumor cells escape apoptosis (35,36). In liver cancer, it was found that BIRC5 is closely related to the functions of promoting cancer cell proliferation, metastasis, inhibiting apoptosis, and participating in angiogenesis (37-39). In our study, 2 active ingredients, namely UA and quercetin, were found to act on BIRC5 protein targets. Ursolic acid (UA), also known as 3beta-hydroxyurs-12-en-28-oic acid, is a natural pentacyclic terpenoid with broad anti-cancer activity (40,41). Previous studies have discovered that UA can promote the apoptosis of liver cancer cells by activating caspase-3 and blocking the PI3K/Akt/survivin signaling pathway (42-44). At the same time, much literature has shown that quercetin can also down-regulate the protein expression of survivin and Bcl-2 in HepG2 cells and induce apoptosis of liver cancer cells (45-46). It can be seen that our research results (43-46) are consistent with previous reports in that UA and quercetin can act on BIRC5 to inhibit the malignant proliferation of HCC.
SPP1, also termed osteopontin (OPN), is an important extracellular glycoprotein in the small integrin-binding ligand N-linked glycoprotein (SIBLING) family (47). Much research has shown that SPP1 is closely linked with processes such as growth, adhesion, invasion, angiogenesis, and metastasis in many malignant tumors (48-50). In liver cancer cells, SPP1 can not only bind to CD44 and activate the PI3K/Akt signaling pathway, but it can also activate integrins and induce the expression of NF-κB to inhibit apoptosis. In addition, SPP1 can also improve the anti-apoptotic ability of HCC by regulating MAPK signaling and binding to EGFR, as well as other related mechanisms (51-54). In our study, it was found that quercetin in the root of ACP can mainly act on the SPP1 target, but the mechanism of action in liver cancer has not been covered so far, and this still needs to be investigated by further experiments. In addition, our study also discovered that the prognostic model established by the 3 genes EGF, BIRC5, and SPP1 was closely associated with the poor prognosis of liver cancer patients, indicating that these 3 genes may be used as early warning markers for liver cancer to a certain extent. Since the 3 target genes EGF, BIRC5, and SPP1 and the corresponding active components of ACP root (quercetin and UA) have critical roles in liver cancer, it is necessary to study the interaction between these target genes and active drug ingredients, as well as their potential link with liver cancer.
More interestingly, related studies (55,56) also found that these ACP root target genes were involved in some immune pathways, which confirmed the potential association between ACP root and the immune system. ACP root has the effect of strengthening the spleen and tonifying qi. It can improve the immune level of the body, strengthen immune surveillance, reverse immune escape, and control tumor growth by regulating spleen deficiency and strengthening the spleen. Therefore, we infer that the root of ACP may achieve anti-HCC efficacy by regulating immune-related factors. As expected, we found that the risk scores of the prognostic models constructed by EGF, BIRC5, and SPP1 were actively correlated with expression levels of different immune cells and common immunosuppression-related genes. Relevant studies at home and abroad have found that EGF, BIRC5, and SPP1 are closely related to the immune escape process in cancer cells. Some scholars have observed that both EGF and SPP1 can up-regulate the expression of PD-L1 in cancer cells and participate in the immunosuppressive process in the cancer cell microenvironment to assist the malignant proliferation of cancer cells (57-60). In addition, related studies have also found that down-regulation of the expression of BIRC5/survivin as a means of immune evasion mediates tumor destruction, thereby affecting the viability of tumor cells (61,62). It can be seen that EGF, BIRC5, and SPP1 play significant roles in mediating the immune escape of tumor cells.
In our research results, it was found that the active components in the root of ACP can regulate the immune escape process of tumor cells and activate the immune system by acting through EGF, BIRC5, and SPP1, which may be potential molecular pathways for the root of ACP in the treatment of liver cancer. The network pharmacology and bioinformatics methods applied in this study mainly rely on a variety of databases, mathematical models and statistical algorithms. Due to the incompleteness of the database, the diversity and complexity of statistical methods and other reasons, our research has limitations to a certain extent. Therefore, we should conduct further research and improvement through a large number of experiments in the future, but this research can provide a certain reference value for basic experiments.
Conclusions
Taken together, our study is the first to identify the potential associations between ACP root, prognostic target genes (EGF, BIRC5, and SPP1), immune regulation, and liver cancer. We speculate that ACP root may inhibit the expression of EGF, BIRC5, and SPP1 target genes, regulate the activity of immune-related factors in the tumor microenvironment, reverse immune escape, stimulate the activity and function of immune cells, enhance immune responses, and exert anti-tumor activity. Our study helps to elucidate the complex mechanism of ACP root against liver cancer and lays a theoretical foundation for further experimental verification.
Acknowledgments
Funding: This project was funded by Scientific Research Project of Shaanxi Provincial Administration of Traditional Chinese Medicine (No. 2019-GJ-JC005), Guiding Plan of Natural Fund of Liaoning Province (No. 2019-ZD-1072), and Shanghai Municipal Health Commission (No. 202040180).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-398/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-398/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen C, Wu B, Zhang C, et al. Immune-related adverse events associated with immune checkpoint inhibitors: An updated comprehensive disproportionality analysis of the FDA adverse event reporting system. Int Immunopharmacol 2021;95:107498. [Crossref] [PubMed]
- Nussinov R, Tsai CJ, Jang H. Anticancer drug resistance: An update and perspective. Drug Resist Updat 2021;59:100796. [Crossref] [PubMed]
- Li JJ, Liang Q, Sun GC. Traditional Chinese medicine for prevention and treatment of hepatocellular carcinoma: A focus on epithelial-mesenchymal transition. J Integr Med 2021;19:469-77. [Crossref] [PubMed]
- Wang K, Chen Q, Shao Y, et al. Anticancer activities of TCM and their active components against tumor metastasis. Biomed Pharmacother 2021;133:111044. [Crossref] [PubMed]
- Gan C, Jin Z, Wei X, et al. Actinidia chinensis Planch. root extract inhibits the proliferation, migration and invasion of breast cancer cells via the AKT/GSK-3β signaling pathway. Folia Histochem Cytobiol 2021;59:226-35. [Crossref] [PubMed]
- Zheng Y, Su L, Tan J, et al. Actinidia chinensis Planch Root extract suppresses the growth and metastasis of hypopharyngeal carcinoma by inhibiting E2F Transcription Factor 1-mediated MNX1 antisense RNA 1. Bioengineered 2022;13:4911-22. [Crossref] [PubMed]
- Yuan C, Wu C, Xue R, et al. Suppression of human colon tumor by EERAC through regulating Notch/DLL4/Hes pathway inhibiting angiogenesis in vivo. J Cancer 2021;12:5914-22. [Crossref] [PubMed]
- Hong Z, Lu Y, Ran C, et al. The bioactive ingredients in Actinidia chinensis Planch. Inhibit liver cancer by inducing apoptosis. J Ethnopharmacol 2021;281:114553. [Crossref] [PubMed]
- Hu W, Wu C, Yuan C, et al. Ethanol Extracted from Radix of Actinidia Chinensis Inhibits Human Colon Tumor Through Inhibiting Notch-signaling Pathway. J Cancer 2021;12:622-9. [Crossref] [PubMed]
- Chen Y, Cai X, Li G, et al. Chemical constituents of radix Actinidia chinensis planch by UPLC-QTOF-MS. Biomed Chromatogr 2021;35:e5103. [Crossref] [PubMed]
- Sun LT, Zhang LY, Shan FY, et al. Jiedu Sangen decoction inhibits chemoresistance to 5-fluorouracil of colorectal cancer cells by suppressing glycolysis via PI3K/AKT/HIF-1α signaling pathway. Chin J Nat Med 2021;19:143-52. [Crossref] [PubMed]
- Fang T, Fang Y, Xu X, et al. Actinidia chinensis Planch root extract attenuates proliferation and metastasis of hepatocellular carcinoma by inhibiting epithelial-mesenchymal transition. J Ethnopharmacol 2019;231:474-85. [Crossref] [PubMed]
- Fang T, Hou J, He M, et al. Actinidia chinensis Planch root extract (acRoots) inhibits hepatocellular carcinoma progression by inhibiting EP3 expression. Cell Biol Toxicol 2016;32:499-511. [Crossref] [PubMed]
- Fang T, Zhao Z, Yuan F, et al. Actinidia Chinensis Planch Root extract attenuates proliferation and metastasis of hepatocellular carcinoma by inhibiting the DLX2/TARBP2/JNK/AKT pathway. J Ethnopharmacol 2020;251:112529. [Crossref] [PubMed]
- Hou J, Wang L, Wu D. The root of Actinidia chinensis inhibits hepatocellular carcinomas cells through LAMB3. Cell Biol Toxicol 2018;34:321-32. [Crossref] [PubMed]
- Zhang X, Qiu H, Li C, et al. The positive role of traditional Chinese medicine as an adjunctive therapy for cancer. Biosci Trends 2021;15:283-98. [Crossref] [PubMed]
- Dong LQ, Peng LH, Ma LJ, et al. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J Hepatol 2020;72:896-908. [Crossref] [PubMed]
- Zongyi Y, Xiaowu L. Immunotherapy for hepatocellular carcinoma. Cancer Lett 2020;470:8-17. [Crossref] [PubMed]
- Fu K, Wang C, Ma C, et al. The Potential Application of Chinese Medicine in Liver Diseases: A New Opportunity. Front Pharmacol 2021;12:771459. [Crossref] [PubMed]
- Chen X, Wang P, Yang M, et al. Therapeutic effect of Jianpi Liqi Fang combined with transcatheter arterial chemoembolization in patients with hepatocellular carcinoma and spleen deficiency syndrome. J Tradit Chin Med 2021;41:157-66. [PubMed]
- Chen F, Li J, Wang H, et al. Anti-Tumor Effects of Chinese Medicine Compounds by Regulating Immune Cells in Microenvironment. Front Oncol 2021;11:746917. [Crossref] [PubMed]
- Zheng J, Xu W, Liu W, et al. Traditional Chinese medicine Bu-Shen-Jian-Pi-Fang attenuates glycolysis and immune escape in clear cell renal cell carcinoma: results based on network pharmacology. Biosci Rep 2021;41:BSR20204421. [Crossref] [PubMed]
- Wu Z, Li S, Zhu X. The Mechanism of Stimulating and Mobilizing the Immune System Enhancing the Anti-Tumor Immunity. Front Immunol 2021;12:682435. [Crossref] [PubMed]
- Shen Q, Zhu J, Jia Y. Analysis on Syndrome and Treatment of Amassment and Accumulation in Jingyue's Complete Works. Journal of Shandong University of Traditional Chinese Medicine 2021;45:191-4.
- Chen H, Wu X, Zhou H, et al. Epidermal growth factor upregulates the expression of A20 in hepatic cells via the MEK1/MSK1/p-p65 (Ser276) signaling pathway. Am J Transl Res 2021;13:708-18. [PubMed]
- Chen H, Li Y, Xiao SY, et al. Identification of a five-immune gene model as an independent prognostic factor in hepatocellular carcinoma. BMC Cancer 2021;21:278. [Crossref] [PubMed]
- Pan Z, Liu C, Zhi Y, et al. LIMK1 nuclear translocation promotes hepatocellular carcinoma progression by increasing p-ERK nuclear shuttling and by activating c-Myc signalling upon EGF stimulation. Oncogene 2021;40:2581-95. [Crossref] [PubMed]
- Chen Y, Cheng L, Yu D, et al. Preparation of Epidermal Growth Factor-Modified Targeted Doxorubicin Nanoliposomes and Therapy of Liver Cancer. J Nanosci Nanotechnol 2021;21:4565-72. [Crossref] [PubMed]
- González L, Díaz ME, Miquet JG, et al. Growth Hormone Modulation of Hepatic Epidermal Growth Factor Receptor Signaling. Trends Endocrinol Metab 2021;32:403-14. [Crossref] [PubMed]
- Zhao X, Wang J, Deng Y, et al. Quercetin as a protective agent for liver diseases: A comprehensive descriptive review of the molecular mechanism. Phytother Res 2021;35:4727-47. [Crossref] [PubMed]
- Liu T, Li Z, Tian F. Quercetin inhibited the proliferation and invasion of hepatoblastoma cells through facilitating SIRT6-medicated FZD4 silence. Hum Exp Toxicol 2021;40:S96-S107. [Crossref] [PubMed]
- Zhang Q, Huang X. The modulatory properties of Astragalus membranaceus treatment on endometrial cancer: an integrated pharmacological method. PeerJ 2021;9:e11995. [Crossref] [PubMed]
- Asgharian P, Tazehkand AP, Soofiyani SR, et al. Quercetin Impact in Pancreatic Cancer: An Overview on Its Therapeutic Effects. Oxid Med Cell Longev 2021;2021:4393266. [Crossref] [PubMed]
- Frazzi R. BIRC3 and BIRC5: multi-faceted inhibitors in cancer. Cell Biosci 2021;11:8. [Crossref] [PubMed]
- Liu X, Zhang H, Zhou P, et al. CREB1 acts via the miR-922/ARID2 axis to enhance malignant behavior of liver cancer cells. Oncol Rep 2021;45:79. [Crossref] [PubMed]
- Wu S, Zang Q, Xing Z, et al. A Pan-Cancer Analysis of the BIRC Gene Family and Its Association with Prognosis, Tumor Microenvironment, and Therapeutic Targets. Crit Rev Eukaryot Gene Expr 2021;31:35-48. [Crossref] [PubMed]
- Mohamed AA, Yassin AS, Gomaa BS, et al. Association of Polymorphism in Survivin Gene and the Risk of Liver Cancer Resulting from Hepatitis C Virus Among Egyptian Patients. Curr Cancer Drug Targets 2021;21:536-43. [Crossref] [PubMed]
- Xu R, Lin L, Zhang B, et al. Identification of prognostic markers for hepatocellular carcinoma based on the epithelial-mesenchymal transition-related gene BIRC5. BMC Cancer 2021;21:687. [Crossref] [PubMed]
- Zhang M, Yan X, Wen P, et al. CircANKRD52 Promotes the Tumorigenesis of Hepatocellular Carcinoma by Sponging miR-497-5p and Upregulating BIRC5 Expression. Cell Transplant 2021;30:9636897211008874. [Crossref] [PubMed]
- Wang L, Yin Q, Liu C, et al. Nanoformulations of Ursolic Acid: A Modern Natural Anticancer Molecule. Front Pharmacol 2021;12:706121. [Crossref] [PubMed]
- Nguyen HN, Ullevig SL, Short JD, et al. Ursolic Acid and Related Analogues: Triterpenoids with Broad Health Benefits. Antioxidants (Basel) 2021;10:1161. [Crossref] [PubMed]
- Ali Abdalla YO, Subramaniam B, Nyamathulla S, et al. Natural Products for Cancer Therapy: A Review of Their Mechanism of Actions and Toxicity in the Past Decade. J Trop Med 2022;2022:5794350. [Crossref] [PubMed]
- Maphanao P, Thanan R, Loilome W, et al. Synchrotron FTIR microspectroscopy revealed apoptosis-induced biomolecular changes of cholangiocarcinoma cells treated with ursolic acid. Biochim Biophys Acta Gen Subj 2020;1864:129708. [Crossref] [PubMed]
- Tang C, Lu YH, Xie JH, et al. Downregulation of survivin and activation of caspase-3 through the PI3K/Akt pathway in ursolic acid-induced HepG2 cell apoptosis. Anticancer Drugs 2009;20:249-58. [Crossref] [PubMed]
- Duan J, Guo H, Fang Y, et al. The mechanisms of wine phenolic compounds for preclinical anticancer therapeutics. Food Nutr Res 2021; [Crossref] [PubMed]
- Tan J, Wang B, Zhu L. Regulation of survivin and Bcl-2 in HepG2 cell apoptosis induced by quercetin. Chem Biodivers 2009;6:1101-10. [Crossref] [PubMed]
- Wei J, Chen Z, Hu M, et al. Characterizing Intercellular Communication of Pan-Cancer Reveals SPP1+ Tumor-Associated Macrophage Expanded in Hypoxia and Promoting Cancer Malignancy Through Single-Cell RNA-Seq Data. Front Cell Dev Biol 2021;9:749210. [Crossref] [PubMed]
- Qian J, LeSavage BL, Hubka KM, et al. Cancer-associated mesothelial cells promote ovarian cancer chemoresistance through paracrine osteopontin signaling. J Clin Invest 2021;131:146186. [Crossref] [PubMed]
- Nallasamy P, Nimmakayala RK, Karmakar S, et al. Pancreatic Tumor Microenvironment Factor Promotes Cancer Stemness via SPP1-CD44 Axis. Gastroenterology 2021;161:1998-2013.e7. [Crossref] [PubMed]
- Pang X, Zhang J, He X, et al. SPP1 Promotes Enzalutamide Resistance and Epithelial-Mesenchymal-Transition Activation in Castration-Resistant Prostate Cancer via PI3K/AKT and ERK1/2 Pathways. Oxid Med Cell Longev 2021;2021:5806602. [Crossref] [PubMed]
- Song Z, Chen W, Athavale D, et al. Osteopontin Takes Center Stage in Chronic Liver Disease. Hepatology 2021;73:1594-608. [Crossref] [PubMed]
- Fnu G, Agrawal P, Kundu GC, et al. Structural Constraint of Osteopontin Facilitates Efficient Binding to CD44. Biomolecules 2021;11:813. [Crossref] [PubMed]
- Lei Y, Yan W, Lin Z, et al. Comprehensive analysis of partial epithelial mesenchymal transition-related genes in hepatocellular carcinoma. J Cell Mol Med 2021;25:448-62. [Crossref] [PubMed]
- Guo DZ, Huang A, Wang YP, et al. Development of an Eight-gene Prognostic Model for Overall Survival Prediction in Patients with Hepatocellular Carcinoma. J Clin Transl Hepatol 2021;9:898-908. [Crossref] [PubMed]
- Park EJ, Kim B, Eo H, et al. Control of IgE and selective T(H)1 and T(H)2 cytokines by PG102 isolated from Actinidia arguta. J Allergy Clin Immunol 2005;116:1151-7. [Crossref] [PubMed]
- Sun HX, Wang H, Xu HS, et al. Novel polysaccharide adjuvant from the roots of Actinidia eriantha with dual Th1 and Th2 potentiating activity. Vaccine 2009;27:3984-91. [Crossref] [PubMed]
- Liu Z, Ning F, Cai Y, et al. The EGFR-P38 MAPK axis up-regulates PD-L1 through miR-675-5p and down-regulates HLA-ABC via hexokinase-2 in hepatocellular carcinoma cells. Cancer Commun (Lond) 2021;41:62-78. [Crossref] [PubMed]
- Suárez GM, Añé-Kourí AL, González A, et al. Associations among cytokines, EGF and lymphocyte subpopulations in patients diagnosed with advanced lung cancer. Cancer Immunol Immunother 2021;70:1735-43. [Crossref] [PubMed]
- Dai M, Yip YY, Todaro G, et al. Antibodies to EGF Receptor Family Members Can Upregulate Tumor Immunity. J Immunother 2021;44:355-61. [Crossref] [PubMed]
- Klement JD, Poschel DB, Lu C, et al. Osteopontin Blockade Immunotherapy Increases Cytotoxic T Lymphocyte Lytic Activity and Suppresses Colon Tumor Progression. Cancers (Basel) 2021;13:1006. [Crossref] [PubMed]
- Xu L, Yu W, Xiao H, et al. BIRC5 is a prognostic biomarker associated with tumor immune cell infiltration. Sci Rep 2021;11:390. [Crossref] [PubMed]
- Andersen MH, Svane IM, Becker JC, et al. The universal character of the tumor-associated antigen survivin. Clin Cancer Res 2007;13:5991-4. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


