
Tripartite motif 52 (TRIM52) promotes proliferation, migration, and regulation of colon cancer cells associated with the NF-κB signaling pathway
Introduction
Recently, the incidence of malignant tumors has been increasing. The latest report from the National Cancer Center states that colorectal cancer (CRC) ranked second in cancer incidence and fifth in cancer mortality in China in 2019 (1). Relevant data from the United States show that the incidence of CRC in men has decreased at a rate of 3% per year over the past 5 years, with no significant change in the incidence rate in women (2). As a cancer with high morbidity and mortality, the mechanism underlying the development of CRC remains unclear and requires further exploration.
The tripartite motif protein (TRIM) family has been found to widely exist in humans (with more than 70 species having been identified) and some multicellular animals. The TRIM protein family is a group of proteins with highly conserved RING, B-box, double helix, and C-terminus (RBCC) structures in order from N-terminal to C-terminal (3). In addition, TRIM proteins are divided into 11 subgroups according to the C-terminal sequence (4,5), among which the PRY/SPRY sequence, also known as the B30.2 sequence, is the most common sequence (5).
A significant portion of the TRIM protein family comprises ubiquitin ligase (E3) activity (6,7), which plays a considerable role in mediating post-translational modifications of the protein by binding to ubiquitin-binding enzymes (E2). E3 activity is closely related to its RING structure. Ubiquitination is a post-translational modification of proteins that is very important in tumorigenesis. Previous studies on the TRIM protein family have shown that TRIM proteins play diverse roles in tumorigenesis. Most of them play a role in promoting tumor progression and metastasis, while others play a tumor-suppressive role (8-10). Certain TRIM family members have been shown to be potential molecular biomarkers for cancer diagnosis and prognosis, and potential therapeutic targets (11).
In mammals, partial gene sequences of Tripartite motif 52 (TRIM52) are duplicated from partial genes of TRIM41, mainly in the RING and B-box structural domains, and have been present in primate mammals for a long time, implying that TRIM52 plays an essential role in certain human biological processes. It has been found that TRIM52 is differentially expressed in different tumor cells. It has been reported that TRIM52 promotes tumorigenesis in ovarian cancer by activating the nuclear factor (NF)-κB signaling pathway. Knocking-down of TRIM52 in ovarian cancer cells suppresses cell invasion, cell migration and cell proliferation, while inducing apoptosis (12). Overexpression of TRIM52 promoted the biological activity of hepatocellular carcinoma. Downregulation of TRIM52 significantly upregulated p21 and PPM1A expression but suppressed the expression of MMP2, which induced dephosphorylation of Smad2/3 in hepatocellular carcinoma cells. In addition, down-regulation of TRIM52 inhibited the ubiquitination of PPM1A. Upregulation of PPM1A in hepatocellular carcinoma cells significantly suppressed the TRIM52-mediated cell biological activity. It can be seen that TRIM52 interacted with PPM1A (13). Downregulation of TRIM52 has been shown to inhibit kidney cancer progression (14). Non-coding (lnc)RNA TRIM52-AS1 knockdown inhibited the proliferation and metastasis of hepatocellular carcinoma cells. It has been shown to promote the growth and metastasis of hepatocellular carcinoma via miR-218-5p/ROBO1 (15). It was reported that TRIM52 upregulation promotes proliferation and invasiveness through activation of the Wnt/βcatenin pathway in lung cancer cells (16). The role that TRIM52 plays in colon cancer (CC) cells remains unknown. CRC is one of the most common cancers around the world as mentioned above. It has been reported that the possible mechanisms by which NF-κB can contribute to colon carcinogenesis include the activator of antiapoptotic gene expression, enhanced cell survival, and proliferation, regulation of angiogenesis, and promotion of metastasis of cancer cells (17). This study aimed to determine the different expression levels of TRIM52 in normal colon epithelial cells and CC cells, explore its effect on CC cells, and identify the underlying mechanisms associated with the NF-κB signaling pathway. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-317/rc).
Methods
Cell lines
We obtained the following cell lines: SW1116 (TCHu174, human colon cancer cells), SW480 (TCHu172, human colon cancer cells), SW620 (TCHu101, human colon cancer cells), LOVO (TCHu82, human colon cancer cells), and HT29 (TCHu103, human colon cancer cells) from the National Collection of Authenticated Cell Culture, Chinese Academy of Sciences (Shanghai, China); HCT116 (human colon cancer cells) and NCM460 (human normal mucosal epithelial cells) from the American-Type Culture Collection (Manassas, VA, USA). Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) at 37 ℃ with 5% CO2. Cells were used in the following assays when they were in the logarithmic growth phase.
Western blot
Protein expressions of TRIM52 and NF-κB signaling pathway associated proteins were tested by western blot (WB). Cell protein was collected from CC cells by radioimmunoprecipitation assay (RIPA) buffer [1 mM with phenylmethylsulfonyl fluoride (PMSF)]. A bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to determine protein concentrations. Lysate samples were separated by electrophoresis on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel at 120v and subsequently transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were incubated with primary antibodies for at least 12 hours at 4 ℃. After the primary antibody was incubated, horseradish peroxidase (HRP)-conjugated secondary antibody was incubated with the membranes for 2 hours in a room temperature environment. Signals were detected using chemiluminescent substrate (ECL; Bio-Rad, Richmond, CA, USA). The band intensity was measured by ImageJ software (National Institutes of Health, Bethesda, MD, USA). Primary antibodies NF-κB P65 (ab32536), phosphor-NF-κB P65 (ab76302), IKKβ (ab32135), IKKα (ab32041), IKBα (ab32518), phosphor-IKBα (ab133462), and β-actin (ab8226) were purchased from Abcam (Cambridge, MA, USA). Antibodies against TRIM52 (sc-398954) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; G9545) served as an internal control and was obtained from Sigma Aldrich (St. Louis, MO, USA). Phosphor-IKKα/β (Ser180/181) (abs130641) was obtained from Absin Bioscience Inc. (Shanghai, China).
Cell migration and invasion assays
Transwell experiments were employed for the cell migration and invasion assays. The upper chamber of the Transwell (Corning, Corning, NY, USA) was coated with Matrigel (Becton, Dickinson, and Co. Biosciences, Franklin Lakes, NJ, USA) at 37 ℃ in a 5% CO2 incubator for 30 minutes for the cell invasion assay. Indicated cells were trypsinized, resuspended in DMEM without serum, and placed in the upper chamber (5×104 cells/well). After that, DMEM with 10% FBS was added to the lower chamber. After 24 hours, a cotton swab was used to remove the cells in the upper chamber. Cells migrating into the lower chamber were washed by the phosphate-buffered saline (PBS), fixed in the 4% paraformaldehyde, and stained by the 0.5% crystal violet. Eventually, the cells were counted under a microscope in 4 random fields. The conditions for the cell migration assay were the same as the cell invasion assay, except for the Matrigel.
Flow cytometry for cell apoptosis
After the transfection for 48 hours, cells were harvested and washed with PBS. Then, the cells were stained with both Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) in sequence at 4 ℃ for 30 minutes in a light-proof environment. Flow cytometry data were collected on the FACScan instrument (Becton Dickinson and Co., Mountain View, CA, USA).
Cell proliferation assay
Cell proliferation assay was detected using a Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. Each assay was performed 6 times.
NF-κB activation assays
Lipopolysaccharide (LPS; an activator of NF-κB P65), and pyrrolidine dithiocarbamate (PDTC; an inhibitor of NF-κB P65) were employed. The LPS (Sigma Aldrich, USA) was co-incubated with the knocked-down TRIM52 HCT116. The PDTC (MedChemExpress, Princeton, NJ, USA) was added to the overexpressed TRIM52 SW620. Protein expressions of nuclear P65 and plasmin P65 were detected to verify the activation of the NF-κB signaling pathway.
Statistical analysis
Data were acquired from at least 3 independent experiments. Statistics were performed in GraphPad Prism7 (GraphPad Software Inc., San Diego, CA, USA). Statistical significance was considered when P<0.05.
Knock down and overexpression of TRIM52 in CC cells
The small interfering (si)RNA (RiboBio, Guangzhou, China) (position 756–774, AGAAATACTGGAAGCATAC, Si-TRIM52 002; position 975–993, GGGCATGTGCTTTAAACAC, Si-TRIM52 003) was added to the HCT116 in the logarithmic growth phase according to the manufacturer’s instructions for 24 hours. The knocking down effect was verified by WB. The coding sequence of TRIM52 was amplified by reverse transcription polymerase chain reaction (RT-PCR) with the following primers: (forward 5'-CGGAATTCATGGCTGGTTATGCCACTACTC-3' and reverse 5'-CGGGATCCTTACTGATTATAGGCCTTGCTG-3'), and constructed into pCDH-CMV-MCS-EF1-CopGFP-T2A-puro (Sygenta Biotechnology Inc., Durham, NC, USA). Lentiviral empty vector was used as a control group. The TRIM52 overexpression lentivirus and control (Vector) lentiviruses were transfected by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
TRIM52 protein expression is up-regulated in CC cells
The CC cells HCT116, SW620, HT29, SW1116, LOVO, and SW480, and the normal colon epithelial cells, NCM460, were cultured, and the expression of TRIM52 was detected after extracting total cell protein by WB. Among CC cells, the expression in HCT116 was the highest, in order of SW480, HT29, SW1116, and LOVO. The expression of TRIM52 was lowest in SW620, which was about 1.6 times more than that in normal colon epithelial cells NCM460 (Figure 1).
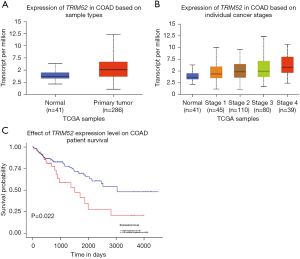
TRIM52 is upregulated in tumor tissues analyzed by TCGA database
The University of Alabama Cancer Database (UALCAN; http://ualcan.path.uab.edu/index.html) was used to find relevant data on TRIM52 expression in CC, and data analysis was performed to speculate on the possible role of TRIM52 in CC. By summarizing TCGA and UALCAN data, it was found that TRIM52 was highly expressed in tumor tissues compared with normal tissues (Figure 2A), and TRIM52 was significantly highly expressed in stage IV tumor tissues, which was statistically different compared with other stages (Figure 2B). This indicated that TRIM52 might be a marker of poorer prognosis. It was also found that the expression of TRIM52 correlated with the survival of tumor patients, and by comparing the survival curves, there was a statistical difference between the high expression TRIM52 group and the low expression group, indicating that TRIM52 may be related to the development of CC and is an indicator of poorer prognosis (Figure 2C).
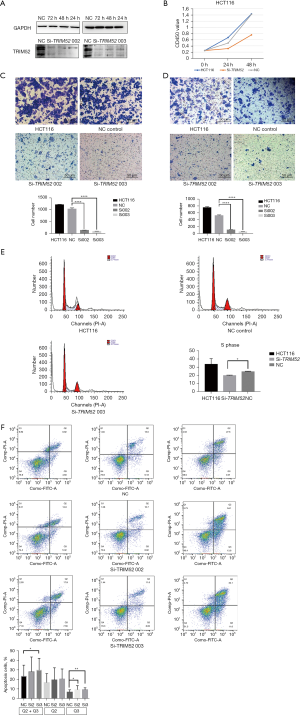
Knockdown of TRIM52 suppresses cell proliferation, migration, and regulation in CC cells
The knockdown effect of siRNA on TRIM52 in HCT116 started to show at 24 hours after transfection and lasted at least until 72 hours (Figure 3A). After knocking down the expression of TRIM52 in HCT116 cells, the proliferation (Figure 3B), invasion (Figure 3C), and migration (Figure 3D) were significantly decreased compared with the untreated HCT116 cell group and the NC control groups. These results showed that the percentage of cells in the S phase decreased from 24.89% to 19.03% after the knockdown of TRIM52 compared to the NC control group, with a statistically significant decrease in the percentage of cells in the S phase (Figure 3E). It was suggested that cellular DNA replication was reduced. Knocking down the expression of TRIM52 in HCT116 cells promoted cell apoptosis and the percentage of late apoptotic cells increased by about 30% (Figure 3F).
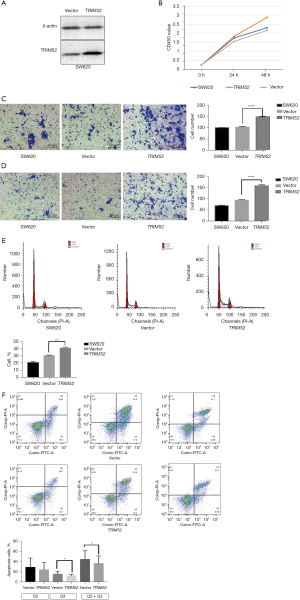
Overexpression of TRIM52 suppresses CRC cell proliferation, migration, and regulation in CC cells
The TRIM52/pCDH-CMV-MCS-EF1-CopGFP-T2A-puro plasmid was more effective for overexpression of TRIM52 in SW620, and its overexpression effect started to show at 24 hours after transfection (Figure 4A). The proliferation (Figure 4B), invasion (Figure 4C), and migration (Figure 4D) ability of SW620 were significantly enhanced after overexpression of TRIM52 by plasmid compared to untreated SW620. The percentage of S-phase cells increased from 30.01% to 40.31% after overexpression of TRIM52 in SW620, which was a statistically significant, suggesting an increase in cellular DNA replication (Figure 4E). Apoptosis was significantly inhibited, and the percentage of early and late apoptosis was reduced by about 20%, which was statistically different from the percentage of late apoptosis after the overexpression of TRIM52 in SW620 (Figure 4F).
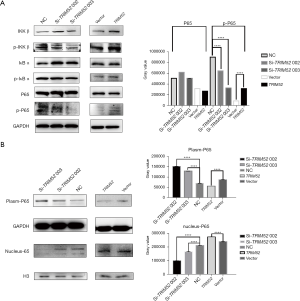
TRIM52 is present in CC cells as a factor that promotes NF-κB activation
By knocking down the expression of TRIM52 in HCT116, the expression of p-IKKβ was decreased, and while the expression of p-IKB was slightly decreased, the expression of P-P65 was decreased significantly both in SiR-TRIM52 002 and SiR-TRIM52 003 group (the gray value of P-P65: NC =9.01×105±41.28, SiR-TRIM52 002=6.45×105±88.49, SiR-TRIM52 003=3.27×105±126.7, n=3, P<0.0001). On the contrary, overexpression of TRIM52 protein in SW620 resulted in the slightly elevated expression of IKKβ, the increased expression of p-IKKβ, the increased expression of p-IKB, and the relatively increased expression of P-P65 (the gray value of P-P65: Vector =8.86×104±8.65, TRIM52=3.17×105±95.35, n=3, P<0.0001) (Figure 5A). Transport of P65 from cytoplasm to nucleus is a characteristic change of the NF-κB signaling pathway activation. It is generally believed that phosphorylated P65, the active form of P65, is present in the nucleus. The results illustrate that knocking down the expression of TRIM52 in HCT116 decreased the expression of p-P65 and the NF-κB signaling pathway was inhibited, while the overexpression of TRIM52 promoted the NF-κB signal pathway activation (Figure 5A). The proteins in the cytoplasm and nucleus were further extracted and examined by WB. It was found that knockdown of TRIM52 expression in HCT116 increased the expression of P65 in the cytoplasm and decreased the expression of P65 in the nucleus (the gray value of nucleus-P65: NC =2.11×105±89.9, SiR-TRIM52 002=9.97×104±68.04, SiR-TRIM52 003=1.66×105±314.2, n=3, P<0.0001). This indicated that the transfer of P65 from the cytoplasm to the nucleus was blocked, and the activation of the NF-κB signaling pathway was inhibited, while overexpression of TRIM52 in SW620 resulted in the opposite (the gray value of nucleus-P65: Vector =2.40×105±89.26, TRIM52=2.74×105±402.6, n=3, P<0.0001) (Figure 5B). Therefore, the presence of TRIM52 worked as a promoter of the NF-κB signal pathway in CC tumor cells.
TRIM52 is closely related to the NF-κB signaling pathway and acts as an agonist in the NF-κB signaling pathway, with its site of action in the upstream pathway of NF-κB complex activation
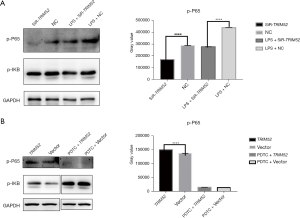
It has been reported that LPS binding to Toll-like receptor 4 (TLR4) causes activation of NF-κB (18). LPS was known as an activator of the NF-κB signaling pathway by promoting the phosphorylation of IKB. After LPS treatment with HCT116, p-IKB was found to be reduced and p-IKB ubiquitination hydrolysis was stimulated, which promoted activation of the NF-κB signaling pathway. The knockdown of TRIM52 expression in the HCT116 cell line still significantly reduced the expression of p-P65 (the gray value of P-P65: NC =2.89×105±95.4, SiR-TRIM52 =1.69×105±51.49, n=3, P<0.0001) and slightly reduced the expression of p-IKB (the gray value of p-IKB: LPS+NC =4.40×105±87.55, LPS+SiR-TRIM52 =2.79×105±89.27, n=3, P<0.0001), indicating that TRIM52 still had an inhibitory effect on the NF-κB signaling pathway after LPS activation, and the site where TRIM52 inhibited the NF-κB signaling pathway acted upstream of the phosphorylation of IKB. In contrast, after inhibition of NF-κB complex activity by PDTC, overexpression of TRIM52 in the SW620 cell line had little effect on the rise of p-P65 expression, and p-P65 was significantly reduced and almost none was expressed (Figure 6A,6B).
Discussion
The TRIM52 protein belongs to the C-V isoform of the TRIM protein family, with 1 RING structure at a time from the N-terminal to the C-terminal, a B-Box2 structural domain, and lacking a distinct double helix structural domain and C-terminus. It has been shown as a promoter in ovarian cancer (12), hepatocellular carcinoma (13,15), and renal cancer (14), while its role in the development of CC had not been clarified
Our study demonstrated that TRIM52 is significantly highly expressed in CC cell lines. Cell proliferation and cell cycle showed a statistically significant decrease of approximately 20.83% in the proportion of S-phase cells in HCT116 cells following knockdown of TRIM52. In contrast, when TRIM52 was overexpressed in SW620, its G2M phase cells increased by about 29.03%. This result fully illustrated that TRIM52 plays a role in the cell cycle to promote cellular DNA replication, and most cells start DNA replication and enter the cell division process after DNA formation of 4-ploidy. The cell cycle results showed that there was a slight increase in the ratio of cells in the G2M phase (the second peak), with no significant statistical difference, which may be related to factors such as the inactivation of key proteins associated with cell division.
In addition, TRIM52 promoted the proliferation of CC cells and accelerated the cell cycle process, which may be related to the promotion of DNA replication in CC cells. In terms of cell migration and invasion, knock-down of TRIM52 expression in the HCT116 cells significantly reduced cell migration and invasion, while overexpression of TRIM52 protein in SW620 enhanced its migration ability. The above results suggested that TRIM52 plays a role in promoting cell migration and invasion in HCT116 and SW620 CC cells, which is closely related to the metastasis of tumors.
The results of apoptosis showed that expression of TRIM52 significantly changed (about 20–30%) in the proportion of apoptotic cells in CC cell lines HCT116 and SW620 in the middle and late stages, and exerted an inhibitory role on the cells. It was also confirmed that the presence of TRIM52 served as a promoting factor of tumor development in CC cell lines HCT116 and SW620. It was shown that TRIM52 promoted cell proliferation by facilitating the DNA replication process in CC cells and promoting cell progression from pre-division to the division phase. Apoptosis experiments demonstrated that TRIM52 acts as an inhibitor of apoptosis in CC cells.
It is widely accepted that activation of the NF-κB signaling pathway promotes tumor cell proliferation, metastasis, and inhibits apoptosis of tumor cells (19-22). Phosphorylation and ubiquitination of IKB are important events in the activation of the NF-κB signaling pathway. TRIM family is closely associated with the activation of the NF-κB signaling pathway due to its conserved RBCC structure, especially the RING structure, which gives it E3 ubiquitin ligase activity (23-25). The regulation of the TRIMs for the NF-κB signaling pathway is diverse, and some members of the triple structural domain protein family are involved in the activation of the NF-κB signaling pathway, for example, TRIM1 is involved in the tumor necrosis factor (TNF) α/LPS-induced activation of the NF-κB signaling pathway (26), TRIM52 acts as a positive regulator to regulate the NF-κB signaling pathway (27), and TRIM22 also has an activating effect on the NF-κB signaling pathway (28). It has been reported that TRIM22/activates NF-κB signaling in glioblastoma by accelerating the degradation of IκBα (29); TRIM47 activates NF-κB signaling via PKC-ε/PKD3 stabilization and contributes to endocrine therapy resistance in breast cancer (30), and TRIM10 activates the NF-κB signaling pathway in osteosarcoma (31). In contrast, some members of the triple structural domain protein family down-regulate the NF-κB signaling pathway in some tumors, namely, TRIM39 inhibits the NF-κB signaling pathway by stabilizing actin expression (32), and TRIM45 inhibits tumor cell proliferation by suppressing the NF-κB signaling pathway (33). The highly conserved RBCC structure of the TRIM protein family, an E3 ubiquitin ligase, characterizes the relationship between TRIM52 and the NF-κB signaling pathway in tumor cells. After knocking down the TRIM52 protein expression in HCT116 tumor cells, p-IKKβ expression was reduced and p-IKB expression was slightly decreased. Also, p-P65 and intracellular P65 were found to be reduced. The result demonstrated the inhibition of the NF-κB signaling pathway from qualitative and localization perspectives. In contrast, the expression of p-IKKβ and p-IKB increased in the SW620 cells overexpressing TRIM52. The expression of P-P65 and intranuclear P65 increased, which indicated that the NF-κB signaling pathway was activated after overexpression of TRIM52 protein in tumor cells, and it was inferred that TRIM52 was closely related to the activation of the NF-κB signaling pathway. Further experiments revealed that the inhibitory effect of knocking down TRIM52 on the NF-κB signaling pathway in tumor cells persisted after activation of the NF-κB signaling pathway by LPS. As an upstream activator of NF-κB, LPS was seen to be an upstream promoter of TRIM52 for the NF-κB signaling pathway. Meanwhile, after PDTC inhibited the NF-κB complex activity, the activation effect of TRIM52 for the NF-κB signaling pathway in the SW620 cell line was not obvious. It can also be inferred that TRIM52, as an upstream stimulator of the NF-κB signaling pathway, lost its stimulating effect via blockade from the downstream PDTC. It was suggested that TRIM52 is closely related to the NF-κB signaling pathway in CC tumor cells and acts as an upstream stimulator of the NF-κB signaling pathway to stimulate the transfer of P65 into the nucleus to activate the NF-κB signaling pathway.
In vivo experiments can be employed in the future to further explore the role of TRIM52 in colon carcinogenesis. In addition, the specific binding sites of TRIM52 require verification by further experiments.
Conclusions
Our study demonstrated that TRIM52 was significantly overexpressed, promoting proliferation, migration, and invasion while inhibiting apoptosis in CC cells. Our study also revealed that the regulation effect of TRIM52 on CC cells is closely related to the NF-κB signaling pathway, and TRIM52 serves as an upstream factor stimulating the transfer of P65 into the nucleus to activate the NF-κB signaling pathway, which provides a potential target for prognosis prediction or treatment of CC.
Acknowledgments
The authors would like to thank Professor Jiang Jianhai’s team from the Biochemistry Laboratory of Fudan Medical College for their guidance and support.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-317/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-317/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-317/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wei W, Zeng H, Zheng R, et al. Cancer registration in China and its role in cancer prevention and control. Lancet Oncol 2020;21:e342-e9. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Borden KL. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem Cell Biol 1998;76:351-8. [Crossref] [PubMed]
- Short KM, Cox TC. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem 2006;281:8970-80. [Crossref] [PubMed]
- Ozato K, Shin DM, Chang TH, et al. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 2008;8:849-60. [Crossref] [PubMed]
- Venuto S, Merla G. E3 Ubiquitin Ligase TRIM Proteins, Cell Cycle and Mitosis. Cells 2019;8. [Crossref] [PubMed]
- Kato K, Ahmad S, Zhu Z, et al. Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. Mol Cell 2021;81:599-613.e8. [Crossref] [PubMed]
- Marzano F, Caratozzolo MF, Pesole G, et al. TRIM Proteins in Colorectal Cancer: TRIM8 as a Promising Therapeutic Target in Chemo Resistance. Biomedicines 2021;9:241. [Crossref] [PubMed]
- Zhan W, Zhang S. TRIM proteins in lung cancer: Mechanisms, biomarkers and therapeutic targets. Life Sci 2021;268:118985. [Crossref] [PubMed]
- Offermann A, Kang D, Watermann C, et al. Analysis of tripartite motif (TRIM) family gene expression in prostate cancer bone metastases. Carcinogenesis 2021;42:1475-84. [Crossref] [PubMed]
- Zhao G, Liu C, Wen X, et al. The translational values of TRIM family in pan-cancers: From functions and mechanisms to clinics. Pharmacol Ther 2021;227:107881. [Crossref] [PubMed]
- Yang W, Liu L, Li C, et al. TRIM52 plays an oncogenic role in ovarian cancer associated with NF-κB pathway. Cell Death Dis 2018;9:908. [Crossref] [PubMed]
- Zhang Y, Tao R, Wu SS, et al. TRIM52 up-regulation in hepatocellular carcinoma cells promotes proliferation, migration and invasion through the ubiquitination of PPM1A. J Exp Clin Cancer Res 2018;37:116. [Crossref] [PubMed]
- Liu Z, Yan HY, Xia SY, et al. Downregulation of long non-coding RNA TRIM52-AS1 functions as a tumor suppressor in renal cell carcinoma. Mol Med Rep 2016;13:3206-12. [Crossref] [PubMed]
- Liu Y, Wu Y, Liu S, et al. Long Non-Coding RNA TRIM52-AS1 Promotes Growth and Metastasis via miR-218-5p/ROBO1 in Hepatocellular Carcinoma. Cancer Manag Res 2021;13:547-58. [Crossref] [PubMed]
- Mu X, Li H, Zhou L, et al. TRIM52 regulates the proliferation and invasiveness of lung cancer cells via the Wnt/betacatenin pathway. Oncol Rep 2019;41:3325-34. [PubMed]
- Seo GS. The role of NF-kappaB in colon cancer. Korean J Gastroenterol 2011;57:3-7. [Crossref] [PubMed]
- Covert MW, Leung TH, Gaston JE, et al. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science 2005;309:1854-7. [Crossref] [PubMed]
- Beg AA, Sha WC, Bronson RT, et al. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature 1995;376:167-70. [Crossref] [PubMed]
- Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death--a new approach to cancer therapy. J Clin Invest 2005;115:2625-32. [Crossref] [PubMed]
- Tamura S, Narita T, Fujii G, et al. Inhibition of NF-kappaB transcriptional activity enhances fucoxanthinol-induced apoptosis in colorectal cancer cells. Genes Environ 2019;41:1. [Crossref] [PubMed]
- Xie B, Nie S, Hu G, et al. The Involvement of NF-kappaB/Klotho Signaling in Colorectal Cancer Cell Survival and Invasion. Pathol Oncol Res 2019;25:1553-65. [Crossref] [PubMed]
- Urano T, Saito T, Tsukui T, et al. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature 2002;417:871-5. [Crossref] [PubMed]
- Toniato E, Chen XP, Losman J, et al. TRIM8/GERP RING finger protein interacts with SOCS-1. J Biol Chem 2002;277:37315-22. [Crossref] [PubMed]
- Meroni G. Genomics and evolution of the TRIM gene family. Adv Exp Med Biol 2012;770:1-9. [Crossref] [PubMed]
- Kim J, Kim JW, Kim DG, et al. Molecular characterization of Rhodeus uyekii tripartite motif protein 1 (TRIM1) involved in IFN-gamma/LPS-induced NF-kappaB signaling. Fish Shellfish Immunol 2018;79:42-51. [Crossref] [PubMed]
- Fan W, Liu T, Li X, et al. TRIM52: A nuclear TRIM protein that positively regulates the nuclear factor-kappa B signaling pathway. Mol Immunol 2017;82:114-22. [Crossref] [PubMed]
- Yu S, Gao B, Duan Z, et al. Identification of tripartite motif-containing 22 (TRIM22) as a novel NF-kappaB activator. Biochem Biophys Res Commun 2011;410:247-51. [Crossref] [PubMed]
- Ji J, Ding K, Luo T, et al. TRIM22 activates NF-kappaB signaling in glioblastoma by accelerating the degradation of IkappaBalpha. Cell Death Differ 2021;28:367-81. [Crossref] [PubMed]
- Azuma K, Ikeda K, Suzuki T, et al. TRIM47 activates NF-kappaB signaling via PKC-epsilon/PKD3 stabilization and contributes to endocrine therapy resistance in breast cancer. Proc Natl Acad Sci U S A 2021;118:e2100784118. [Crossref] [PubMed]
- Xi X, Bao Y, Zhou Y, et al. Oncogenic gene TRIM10 confers resistance to cisplatin in osteosarcoma cells and activates the NF-kappaB signaling pathway. Cell Biol Int 2021;45:74-82. [Crossref] [PubMed]
- Suzuki M, Watanabe M, Nakamaru Y, et al. TRIM39 negatively regulates the NFkappaB-mediated signaling pathway through stabilization of Cactin. Cell Mol Life Sci 2016;73:1085-101. [Crossref] [PubMed]
- Shibata M, Sato T, Nukiwa R, et al. TRIM45 negatively regulates NF-kappaB-mediated transcription and suppresses cell proliferation. Biochem Biophys Res Commun 2012;423:104-9. [Crossref] [PubMed]



