Chemosensitivity of gastric cancer: analysis of key pathogenic transcription factors
Introduction
Gastric cancer is one of the most common malignant tumors globally and the second leading cause of cancer death (1). At the time of diagnosis, about 30% of patients with gastric cancer have progressed to an advanced stage (2). Chemotherapy is one of the standard treatments for most advanced gastric cancers. However, due to tumor heterogeneity and individual differences, there are significant differences in the sensitivity of different patients to chemotherapy drugs (3). Multidrug resistance of patients to chemotherapy drugs is the main reason that treatment of gastric cancer fails (4). The effect of chemotherapy in patients with gastric cancer depends on chemotherapeutic drugs and is directly or indirectly affected by function-related genes (5). Individual gene changes, or polymorphisms, significantly affect patients’ responses to specific chemotherapy drugs. Therefore, exploring the key pathogenic genes of gastric cancer and their mechanism of action can provide the basis for clinical chemotherapeutic drug selection, promote the development of individualized treatment, and improve the prognosis of gastric cancer patients.
The Genomics of Drug Sensitivity in Cancer (GDSC) database is a collection of data from 75,000 experiments describing the response of approximately 200 anticancer drugs in more than 1,000 tumor cells. The GDSC database can be used to analyze the sensitivity and response of tumor cells to drugs. Variations in the cancer genome can affect the efficacy of clinical treatments, and different targets respond to drugs differently. GDSC data are important for discovering potential tumor therapeutic targets. A study (6) used GDSC database analysis to determine the characteristic gene sets of advanced gastric cancer and the correlation between these gene sets and fluorouracil, which can provide clues for determining the best combination of chemotherapy drugs for gastric cancer patients. Another study (7) constructed a risk score based on gene expression, which was used to predict the survival of gastric cancer patients and the sensitivity to chemotherapy drugs.
In this study, the differentially expressed transcription factors in gastric cancer and normal gastric tissues were screened using the RNA sequencing (RNA-seq) data of gastric cancer in the Cancer Genome Atlas (TCGA) database. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, interaction network construction, and prognostic correlation analysis were used to analyze the differentially expressed transcription factors. The interaction network and prognostic correlation analysis determined FOXM1 as a critical gene. We used GDSC to explore the correlation between FOXM1 gene expression and gene mutation and chemosensitivity. We present the following article in accordance with the STREGA reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-274/rc).
Methods
Data download
RNA-seq data of gastric cancer were downloaded from TCGA, which is public data (normal tissue =32 cases, gastric cancer tissue =375 cases). The downloaded RNA-seq data were combined into a gene expression matrix, and the matrix was normalized and logarithmically transformed. Tumor-related transcription factors were obtained according to Cistrome Project, and their expression was extracted from the data matrix to obtain a new matrix. The row name of the matrix was set as the sample serial number, and the column name was set as the expression of transcription factors. Single nucleotide polymorphism (SNP) data were downloaded from the TCGA database to obtain the mutation status of transcription factors in each sample. The corresponding clinical data of gastric cancer patients were downloaded from the TCGA database, including patient age, gender, survival status, and survival time. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differential expression analysis of transcription factors
The differential expression of transcription factors between gastric cancer and normal tissues was analyzed with the edgeR package. Fold change (FC) was the ratio of expression in gastric cancer tissue to that in normal tissue. In this study, the transcription factors differentially expressed in normal tissues and gastric cancer were screened under the screening conditions of | log2FC | ≥1 and false discovery rate (FDR) <0.05.
Pathway enrichment analysis of KEGG
The KEGG database was used to analyze the pathway enrichment of differentially expressed transcription factors. FDR <0.05 was used as the screening standard to screen the pathways with significant differences.
Screening of prognostic transcription factors
The differentially expressed transcription factors were used as potential prognostic factors for univariate Cox analysis by combining the expression of differentially expressed transcription factors with the overall survival status and overall survival time in the patient’s clinical information. P<0.05 was used as the screening standard to screen the transcription factors related to the prognosis of patients with gastric cancer.
Construction of transcription factor interaction network
The interaction network between differentially expressed transcription factors was constructed with the String database. Key genes were determined according to the number of interactions between transcription factors.
Correlation between mutation and expression
Spearman test was used to explore the correlation between transcription factor mutation and gene expression.
Correlation between expression of transcription factors and chemosensitivity
GDSC database was used to explore the markers for predicting chemosensitivity. According to the expression of key genes in cancer cells, cancer cells were divided into high expression group and low expression group, and the half maximal inhibitory concentration (IC50) of high and low expression groups against different chemotherapeutic drugs was compared.
Statistical analysis
All data were statistically analyzed by R software (MathSoft, USA) and related R package in this study. P<0.05 indicates that it is statistically significant.
Results
Differential transcription factor
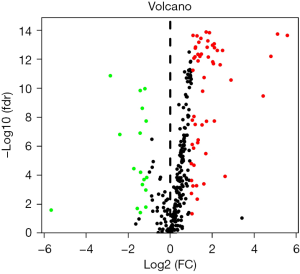
Taking | log2FC | ≥1 and FDR <0.05 as the screening conditions, the expression of transcription factors in 32 normal tissues and 375 gastric cancer tissues were compared. A total of 17 differentially expressed transcription factors were screened. Compared with normal tissues, 15 transcription factors were upregulated and 2 transcription factors were downregulated in gastric cancer tissues, as shown in Table 1. The volcano diagram of differentially expressed transcription factors is shown in Figure 1.
Table 1
| Gene | Normal | Tumor | Log2FC | FDR |
|---|---|---|---|---|
| RXRG | 0.97 | 0.14 | −2.85 | 1.38E-11 |
| MYH11 | 622.04 | 117.87 | −2.40 | 1.55E-07 |
| SOX9 | 17.15 | 69.19 | 2.01 | 1.07E-13 |
| CENPA | 1.28 | 5.18 | 2.02 | 1.50E-12 |
| NCAPG | 1.22 | 5.07 | 2.06 | 2.00E-12 |
| SOX4 | 8.33 | 35.63 | 2.10 | 8.69E-14 |
| MYB | 1.35 | 5.79 | 2.10 | 1.78E-08 |
| E2F1 | 2.58 | 11.11 | 2.11 | 1.61E-13 |
| FOXM1 | 3.59 | 16.97 | 2.24 | 2.46E-13 |
| MYBL2 | 7.70 | 39.94 | 2.38 | 2.44E-12 |
| E2F7 | 0.35 | 1.96 | 2.49 | 2.48E-13 |
| EMX1 | 0.14 | 0.85 | 2.61 | 2.48E-13 |
| CBX2 | 0.36 | 2.70 | 2.89 | 2.66E-11 |
| HOXA9 | 0.08 | 1.66 | 4.42 | 3.38E-10 |
| HOXC11 | 0.09 | 2.37 | 4.79 | 6.18E-13 |
| HOXC9 | 0.07 | 2.31 | 5.11 | 1.86E-14 |
| SALL4 | 0.05 | 2.15 | 5.57 | 2.23E-14 |
FC, fold change; FDR, false discovery rate.
Enrichment analysis of differentially expressed transcription factor in the KEGG pathway
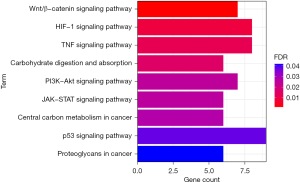
The differentially expressed transcription factors were enriched and analyzed with the KEGG database. Differential transcription factors were significantly enriched in Wnt/β-catenin signaling pathway, HIF-1 signaling pathway, TNF signaling pathway, central carbon metabolism in cancer, PI3K-Akt signaling pathway, JAK-STAT signaling pathway, carbohydrate digestion and absorption, p53 signaling pathway, and proteoglycans in cancer (Figure 2).
Prognosis related genes
This study combined the expression of differentially expressed transcription factors with the overall survival status and overall survival time in patients’ clinical information. It used differentially expressed transcription factors as potential prognostic factors for univariate Cox analysis. Taking P<0.05 as the screening criterion, a total of 8 transcription factors were prognostic factors. Among them, CENPA, E2F1, EMX1, HOXA9, FOXM1, and MYBL2 were prognostic risk factors (HR >1; P<0.05), and RXRG and SOX4 were prognostic protective factors (HR <1; P<0.05; Table 2).
Table 2
| ID | HR | 95% CI | P value | |
|---|---|---|---|---|
| Lower | Upper | |||
| CENPA | 1.42 | 1.04 | 2.18 | 0.002 |
| E2F1 | 2.16 | 1.08 | 3.14 | 0.003 |
| EMX1 | 1.89 | 0.93 | 2.33 | 0.014 |
| HOXA9 | 2.79 | 1.65 | 3.47 | 0.026 |
| FOXM1 | 2.46 | 1.4 | 3.15 | 0.037 |
| MYBL2 | 1.01 | 0.83 | 1.26 | 0.041 |
| RXRG | 0.88 | 0.51 | 1.14 | 0.001 |
| SOX4 | 0.67 | 0.49 | 0.94 | 0.021 |
HR, hazard ratio; CI, confidence interval.
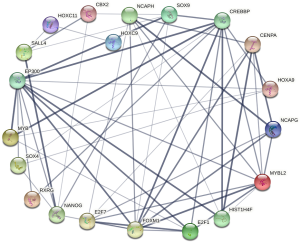
Construction of differentially expressed transcription factor interaction network
Differentially expressed transcription factors in normal tissues and gastric cancer tissues interacted, as shown in Figure 3. FDXM1 interacted with E2F7, MYBL2, E2F1, NCAPG, and SOX9. Using the number of interacting transcription factors as the screening criteria, FOXM1 was the most widely interacted gene in the differentially expressed transcription factor network associated with the prognosis of gastric cancer.
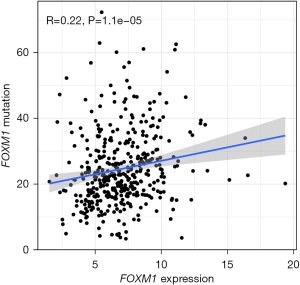
Correlation between FOXM1 transcription factor mutation and expression
Since the above results indicated that FOXM1 was the most widely interacting transcription factor, it was considered as a potential biomarker for gastric cancer, and the relationship between its mutation and expression was further investigated. FOXM1 gene mutation was positively correlated with mRNA expression (R =0.22; P<0.05), as shown in Figure 4.
FOXM1 expression and chemosensitivity
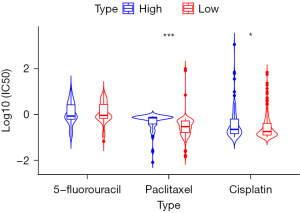
Based on the median value of FOXM1, the gastric cancer cells were divided into FOXM1 high-expression group and low-expression group. There was no significant difference in IC50 of 5-fluorouracil between the FOXM1 high expression group and the low expression group (P>0.05). The IC50 of paclitaxel in the FOXM1 high expression group was significantly higher than that in the FOXM1 low expression group (P<0.001). The IC50 of cisplatin in the FOXM1 high expression group was significantly higher than in the FOXM1 expression group (P<0.05; Figure 5).
Discussion
The mechanism of drug resistance in gastric cancer is very complex. Mechanisms such as intracellular drug efflux and intracellular drug redistribution, changes in the level of intracellular drug-targeted enzymes, and enhanced cellular repair DNA damage can enhance the drug resistance of gastric cancer cells. Changes in the tumor microenvironment and the expression of therapeutic targets may play a key role in the drug resistance of gastric cancer. It is currently believed that some genes may be associated with drug resistance in gastric cancer, including MDR1, MRP1, mTOR and HIF-1α (8). Exploring the relationship between the expression of key pathogenic genes in gastric cancer and the sensitivity to chemotherapeutic drugs can avoid gastric cancer drug resistance and provide a basis for clinical chemotherapeutic drug selection.
Our results showed that the differentially expressed transcription factors were significantly enriched in the gene pathway of the Wnt/β-catenin signaling pathway, the PI3K-Akt signaling pathway, and the p53 signaling pathway, which are a common cancer pathogenic signaling pathway. Existing research confirmed that miRNA, mRNA and lncRNA promote or inhibit tumor progression by activating or inhibiting these pathways (9-14). Oliveira et al. (9) compared the difference of protein expression on the Wnt pathway in 72 gastric cancer specimens with the immunohistochemical method. This study confirmed that Wnt was involved in the progression of gastric cancer. Singh et al. (10) indicated that the PI3K-Akt signaling pathway is frequently activated in gastric cancer. The imbalance of this pathway leads to the occurrence of gastric cancer. Therefore, targeted treatment is needed to obtain more effective anticancer therapy. Singh et al. (10) also used PI3K-Akt signaling pathway to predict biomarkers and promoted personalized cancer treatment and effectively inhibiting the PI3K-Akt signaling pathway. The p53 signaling pathway is a common cancer pathogenic pathway (11,12), and its role in gastric cancer has also been widely confirmed (13,14). Our pathway enrichment analysis results demonstrated that the differentially expressed transcription factors we screened may play a key role in the occurrence and progression of gastric cancer.
FOXM1 was the key transcription factor of gastric cancer identified in this study. We found that FOXM1 was highly expressed in gastric cancer, and its gene expression was positively correlated with gene mutation. We also found that FOXM1 was a risk factor for the prognosis of patients with gastric cancer. These results suggest that FOXM1 may promote the occurrence and progression of gastric cancer. A study pointed out that FOXM1 is overexpressed in Pan-cancer, which is related to the occurrence and progression of most malignant tumors (15). The study showed that the expression of FOXM1 was significantly increased at mRNA and protein levels in tumors with FOXM1 mutation, p53 inactivation and Rb-E2F abnormality (15). The results also illustrated that E2F and cyclin E1 regulate FOXM1 (15). It is important to point out that Barger et al. (15) found a mutation in FOXM1 in a pan-cancer study, resulting in a significant increase in FOXM1 expression, which is consistent with our conclusion. Furthermore, Wierstra (16) suggested that transcription factor FOXM1 is overexpressed in many human cancers, including liver cancer, gastric cancer, prostate cancer, brain cancer, breast cancer, lung cancer, colon cancer, pancreatic cancer, cervical cancer, ovarian cancer, and nervous system cancer. FOXM1 was involved in cell initiation, progression, metastasis, and the response to anti-cancer drugs.
We found that the IC50 of paclitaxel and cisplatin in the FOXM1 high expression group was higher than that in the FOXM1 low expression group. The results indicated that the higher the expression of FOXM1, the less sensitive the patients were to paclitaxel and cisplatin. There was no significant difference in the IC50 of FOXM1 high expression group and low expression group to 5-fluorouracil. Our analysis suggested that FOXM1 may not be involved in the induction of gastric cancer sensitivity to all chemotherapeutic drugs. Some existing studies also showed the relationship between FOXM1 and chemoresistance, which is consistent with our results (17-20). For example, Okada et al. (17) identified 53 gastric cancer patients whose immunohistochemistry was FOXM1 positive. Okada et al. (17) also found that FOXM1 expression was an important independent prognostic risk factor for overall survival and disease-free survival in gastric cancer patients through multivariate analysis. The results of Okada et al. (17) also showed that FOXM1 overexpression was significantly correlated with the drug resistance of docetaxel + 5-fluorouracil + cisplatin chemotherapy. In contrast, the 5-fluorouracil + cisplatin chemotherapy drug resistance was not significant in patients with advanced gastric cancer. Inhibiting overexpression of FOXM1 was a promising strategy for the treatment of advanced gastric cancer. Li et al. (18) considered that FOXM1 overexpression mediates the resistance of gastric cancer to docetaxel. They showed that FOXM1 acts with the downstream target tubulin unstable protein stathmin to change microtubule dynamics to protect tumor cells from docetaxel-induced apoptosis. Their immunohistochemical analysis showed a correlation between the expression levels of FOXM1 and stathmin in 103 postoperative specimens of gastric cancer. Li et al. (18) also found that, when FOXM1 inhibitor sulfur chain protein was used to reduce the expression of FOXM1, the resistance of gastric cancer to docetaxel was reversed, and FOXM1 and stathmin were downregulated. Therefore, FOXM1 can be used as a useful marker for predicting and monitoring docetaxel response. Furthermore, docetaxel resistance can be reversed by inhibiting FOXM1. FOXM1 may become a new therapeutic target for docetaxel-resistant gastric cancer. In other cancers, high expression of FOXM1 also leads to increased resistance to chemotherapeutic drugs. Wang et al. (19) found that FOXM1 expression was significantly correlated with cisplatin-based chemotherapy resistance and poor prognosis in patients with advanced non-small cell lung cancer. Lin et al. (20) indicated that FOXM1 participates in docetaxel resistance in castration-resistant prostate cancer by inducing AMPK/mTOR mediated autophagy. These findings support our findings. FOXM1 plays an oncogenic role in most cancers and is associated with chemoresistance. However, our study is limited to data analysis from public repositories and requires validation of experimental data in vivo and in vitro.
In conclusion, we found that FOXM1 is a transcription factor that is highly expressed in gastric cancer. The high expression of FOXM1 is associated with paclitaxel and cisplatin resistance in patients with gastric cancer. Therefore, FOXM1 is a potential target for the treatment of gastric cancer.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-274/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-274/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ben Kridis W, Marrekchi G, Mzali R, et al. Prognostic factors in metastatic gastric carcinoma. Exp Oncol 2019;41:173-5. [PubMed]
- Hattori Y, Sentani K, Shinmei S, et al. Clinicopathological significance of RCAN2 production in gastric carcinoma. Histopathology 2019;74:430-42. [Crossref] [PubMed]
- Lai J, Li J, Mo X. Clinical Effectiveness of Neoadjuvant Chemotherapy in Gastric Carcinoma and Exploration of Perioperative Imaging Assessment Parameters. Gastroenterol Res Pract 2021;2021:5563136. [Crossref] [PubMed]
- Li H, Lu H, Lv M, et al. Parthenolide facilitates apoptosis and reverses drug-resistance of human gastric carcinoma cells by inhibiting the STAT3 signaling pathway. Oncol Lett 2018;15:3572-9. [Crossref] [PubMed]
- Jin X, Jiang ML, Wu ZH, et al. Progress of Individualized Chemotherapy for Gastric Carcinoma Under the Guidance of Genetic Testing. Curr Med Chem 2020;27:2322-34. [Crossref] [PubMed]
- Lee J H, Park Y R, Jung M, et al. Gene regulatory network analysis with drug sensitivity reveals synergistic effects of combinatory chemotherapy in gastric cancer. Sci Rep 2020;10:3932. [Crossref] [PubMed]
- Jia X, Chen B, Li Z, et al. Identification of a Four-Gene-Based SERM Signature for Prognostic and Drug Sensitivity Prediction in Gastric Cancer. Front Oncol 2022;11:799223. [Crossref] [PubMed]
- Zhang X, Xie K, Zhou H, et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol Cancer 2020;19:47. [Crossref] [PubMed]
- Oliveira LA, Oshima CTF, Soffner PA, et al. The canonical Wnt pathway in gastric carcinoma. Arq Bras Cir Dig 2019;32:e1414. [Crossref] [PubMed]
- Singh SS, Yap WN, Arfuso F, et al. Targeting the PI3K/Akt signaling pathway in gastric carcinoma: A reality for personalized medicine? World J Gastroenterol 2015;21:12261-73. [Crossref] [PubMed]
- Khan H, Reale M, Ullah H, et al. Anti-cancer effects of polyphenols via targeting p53 signaling pathway: updates and future directions. Biotechnol Adv 2020;38:107385. [Crossref] [PubMed]
- Huang J. Current developments of targeting the p53 signaling pathway for cancer treatment. Pharmacol Ther 2021;220:107720. [Crossref] [PubMed]
- Yu YF, Zhang Y, Shen N, et al. Effect of VEGF, P53 and telomerase on angiogenesis of gastric carcinoma tissue. Asian Pac J Trop Med 2014;7:293-6. [Crossref] [PubMed]
- Hou J, Tan Y, Su C, et al. Inhibition of protein FAK enhances 5-FU chemosensitivity to gastric carcinoma via p53 signaling pathways. Comput Struct Biotechnol J 2020;18:125-36. [Crossref] [PubMed]
- Barger CJ, Branick C, Chee L, et al. Pan-Cancer Analyses Reveal Genomic Features of FOXM1 Overexpression in Cancer. Cancers (Basel) 2019;11:251. [Crossref] [PubMed]
- Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res 2013;119:191-419. [Crossref] [PubMed]
- Okada K, Fujiwara Y, Takahashi T, et al. Overexpression of forkhead box M1 transcription factor (FOXM1) is a potential prognostic marker and enhances chemoresistance for docetaxel in gastric cancer. Ann Surg Oncol 2013;20:1035-43. [Crossref] [PubMed]
- Li X, Yao R, Yue L, et al. FOXM1 mediates resistance to docetaxel in gastric cancer via up-regulating Stathmin. J Cell Mol Med 2014;18:811-23. [Crossref] [PubMed]
- Wang Y, Wen L, Zhao SH, et al. FoxM1 expression is significantly associated with cisplatin-based chemotherapy resistance and poor prognosis in advanced non-small cell lung cancer patients. Lung Cancer 2013;79:173-9. [Crossref] [PubMed]
- Lin JZ, Wang WW, Hu TT, et al. FOXM1 contributes to docetaxel resistance in castration-resistant prostate cancer by inducing AMPK/mTOR-mediated autophagy. Cancer Lett 2020;469:481-9. [Crossref] [PubMed]
(English Language Editor: C. Mullens)