
Evaluating prognostic value and relevant gene signatures of tumor microenvironment characterization in esophageal carcinoma
Introduction
Esophageal carcinoma (EC) is one of the most aggressive cancers. It was ranked 9th and 6th for incidence and mortality, respectively (1). The standard therapies for EC are surgery, chemotherapy, and radiotherapy. At present, 3 therapeutic agents that target human epidermal growth factor receptor 2 (HER2), vascular endothelial growth factor 2 (VEGF-2), and programmed cell death protein 1 (PD-1) have been approved by the Food and Drug Administration for use in the treatment of EC (2). Despite the great improvement in therapeutics, the 5-year survival rate remains poor at about 20% (3). Previous genomic studies have explored the molecular mechanism underlying EC, but have mainly focused on the genetic landscape (4,5). The tumor microenvironment (TME), which contains diverse immune cell populations, is thought to play a crucial role in the development of different cancers, including breast cancer (6), gastric cancer, (7) and esophageal squamous cell carcinoma (ESCC) (8), which suggest that immune cell infiltration may be associated with the clinical outcome of cancers. During the development of tumors, oncogenic mutations allow cells to proliferate independently, and cancer cells interact with the TME to favor its expansion and evade immune control (9).
A greater understanding of the tumor-immune microenvironment may provide more advanced prognostic biomarkers for the immunotherapy and chemotherapy of EC patients and reveal additional novel targets. Immunotherapy is a well-known treatment that works by modulating the TME. In addition, research suggests that the success of chemotherapy and radiotherapy relies on the induction of a durable anti-cancer immune response, and that targeted therapies also have immuno-stimulatory effects (10-12). It may be that the TME could change as a result of therapies, such that there is a shift from a pre-existing immune response to a therapy-induced immune response (10). Thus, information about the prediction of a treatment response and prognosis could be gained from the TME. A number of studies have highlighted the role of the TME in prognosis and treatment. For example, a high density of T lymphocytes in TME is associated with a favorable prognosis, while a high density of M2 macrophages is associated with a poor prognosis (10,13,14). The TME context determined at the time of diagnosis could also reflect the immune response and chemotherapy benefits (15,16). PD-L1 blockade is used in the microsatellite instability (MSI)/deficient mismatch repair (dMMR) treatment of colorectal cancers (17). Thus, it is necessary to integrate existing medical and targetable biomarkers, including the MSI biomarker, for the comprehensive evaluation of EC.
Recently, several bioinformatic approaches have been developed to analyze the TME and have been successfully applied to some tumor types (7,18-22). However, little research has been conducted on the use of TME in evaluating the therapeutic efficacy of treatments for patients with EC. In relation to estimations of the TME, immunohistochemistry (IHC) is the only technique to yield quantitative information on the TME. In this study, we analyzed the immune infiltrate of EC by using a proposed computational algorithm, and developed an approach to quantify the TME infiltration pattern (i.e., the TME score). Our results showed that the TME score has potential clinical application and value in evaluating the prognosis of patients with EC. We further showed significant correlations between some micro ribonucleic acids (miRNAs) and differentially expressed genes (DEGs) in TME patterns with survival.
Purpose
This study designed a TME signature to evaluate the comprehensive TME of EC, and a TME score classifier to effectively predict the prognosis of EC patients. The results of this study may have important implications for identifying subgroups of EC patients with a low or high risk of poor survival. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-371).
Methods
Data collection
Molecular data from The Cancer Genome Atlas (TCGA) for EC (ESCA) type, including RNA-sequencing data, single nucleotide variation (SNV) date, copy number variations (CNV) date, miRNA microarray data, methylation microarray data, and corresponding clinical data with survival information, were downloaded (https://portal.gdc.cancer.gov/projects/TCGA-ESCA). Samples without survival information were excluded from further analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data pre-processing
Genes with a low expression level were removed by supplying RNA-sequence raw read count data to the filterByExpr function of edgeR. The read counts of the remaining genes were then normalized by Voom in the Limma package to facilitate the deconvolution of immune cell types.
Estimation of immune cell type fractions in the TME
CIBERSORT is an algorithm for enumerating the immune cell composition of complex tissues from bulk RNA-sequencing data. To infer the fraction of 22 immune cell types in tumor samples, a leukocyte gene signature matrix containing 547 genes (LM22) was used. We uploaded the pre-treated RNA-sequencing data (n=160) to the CIBERSORT web portal (http://cibersort.stanford.edu/), and ran the algorithm using the LM22 gene signature and 1,000 iterations.
Consensus clustering for TME-immune cells
TME clusters were identified using k-means clustering methods in the ConsensuClusterPlus R package with 1,000 resampling times to ensure the stability of classification. The optimal number of clusters was determined using the elbow method. The significance of the survival difference among patients belonging to different TME clusters was assessed by the log-rank test and Kaplan-Meier survival curves. Correlations between immune cell types and survival were calculated by a Cox regression, and the Pearson correlation coefficient (PCC) was used to measure the relationship among different immune cell types.
DEG analysis
DEGs between 2 TME clusters were identified by the Limma package in R with the significance criteria of a P value <0.001 and a |log2FC| >1. To explore the biological implications of these DEGs, functional enrichment was carried out using the R package cluster Profiler.
Generation of TME gene signatures
The signature genes most closely related to the TME clusters were selected by reducing redundant DEGs using a random-forest algorithm. These selected signature genes were then separated into 2 categories according to the correlation between the Cox coefficients and survival. After calculating the coefficient, we defined the TME score, which is similar to the previously reported Gene expression Grade Index (GGI), to summarize the total expression level of survival-related genes. The TME score is calculated according to the following formula:
TME score = Σ voom(X) – Σ voom(Y)
where X is the expression level of genes whose Cox efficient is positive, and Y is the expression level of genes whose Cox efficient is negative.
Exploration of the predictive value of the TME score for prognosis
To explore the relationship between the TME score and the survival of patients, we classified patient into 2 groups based on the optimal breakpoint of TME score determined by R package maxstat. The survival difference between the high and low TME score groups of patients was assessed using the log-rank test and Kaplan-Meier survival curve.
Identification of somatic mutations
SNV data, comprising 160 samples, were used to obtain the mutation spectrum. The maftools R package was used.
Identification of mutational signatures
SNV data, including 160 samples, were used to obtain the mutational spectra and identify mutational signatures. The maftools and SomaticSignatures R packages were used.
Inference of the copy number variation (CNV)
The CNV regions among the 160 samples were detected by the Genomic Identification of Significant Targets in Cancer (GISTIC) module of GenePattern. The confidence interval was set as 0.95, and the other parameters were default values.
Identification of prognosis-related differentially expressed mRNA and miRNA
As stated above, the patients were divided into high and low TME score groups. The differentially expressed messenger RNA (mRNA) and miRNA of these 2 groups were identified using the Limma R package. Next, a gene annotation enrichment analysis was performed. For the analysis of mRNA, the significance criteria were a P<5e-2 and a |log2FC| >0.5. For the analysis of miRNA, the significance criteria were a P<0.05 and a |log2FC| >0.25. For the analysis of methylated sites, the significance criteria were a P<0.05 and a |log2FC| >0.5. Finally, the differentially expressed mRNA and miRNA were further analyzed to explore their correlation with survival.
Statistical analysis
The data analyses were performed with R software (version 3.5.1) and R Bioconductor packages. The normality of the variables was examined using the Shapiro-Wilk normality test. Comparisons between the 2 groups were performed using unpaired Student t-tests if the variables were normally distributed, or otherwise, Wilcoxon rank-sum tests. The survival curves for the subgroups were generated using the Kaplan-Meier method, and the statistical significance of the differences was determined using the log-rank test. A P<0.05 was considered statistically significant.
Data availability
All the data used in this study are available for download from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/projects/TCGA-ESCA).
Results
TME score subtypes as biomarkers for predicting the prognosis of EC
All 160 EC samples with overall survival (OS) information were used for the prognostic model construction. A univariate Cox regression analysis was performed to estimate the prognostic relationship between the TME score and OS in EC patients. Through the LASSO Cox regression model with minimized lambda, the TME score subtypes of 22 immune cell signatures were selected to build the TME score model in the training cohort. The associations between the TME score subtypes of 22 immune cell signatures and OS are presented in Figure 1A. Additionally, the TME cell network depicted tumor-immune cell interactions and their effects on the OS of patients with EC using Cytoscape (see Figure 1B and Table S1). The TME cell network revealed that eosinophils (23), M0 macrophages, M2 macrophages, natural killer cells, and memory cluster of differentiation (CD)4+ T cells (24) were significantly negatively correlated with prognosis in EC, while activated dendritic cells (DCs), CD8+ T cells, and naive CD4+ T cells were significant positively correlated with prognosis in EC. Next, to achieve optimal clustering stability, the unsupervised clustering method (K-means) and elbow method were applied to identify the optimal K value. As a result, 2 clusters were determined (K=2; see Figure S1A, S1B). 2 distinct patterns of TME cell infiltration were determined (K=2) by unsupervised clustering (see Figure S1C), and the proportions of immune cells were slightly different between TME cluster 1 and TME cluster 2 (see Figure S1D).
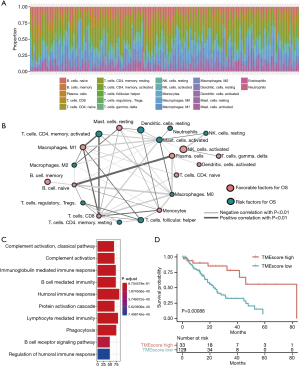
To further understand the biological and clinical differences of the TME phenotypes, the DEGs between TME cluster 1 and TME cluster 2 were determined using the Limma R package. As a result, a total of 687 DEGs were included in the TME classification. An unsupervised clustering analysis was also performed to group patients into genomic subtypes (see Figure S2A, S2B), and the clustering results were significantly consistent with those of the TME phenotype groups (Pearson’s Chi-squared test, P<0.001). We then applied a random-forest algorithm to reduce redundant DEGs, and obtained 119 signature genes (see Tables S2, S3). The biological processes of gene ontology (GO) terms were mainly enriched in immune-related processes, such as the humoral immune response mediated by circulating immunoglobulin, the immunoglobulin-mediated immune response, and B cell-mediated immunity (see Figure 1C). Based on the Cox regression model, the Cox coefficient for each cluster signature gene was obtained and used to calculate TME scores. As a result, the samples were classified into high or low TME scores. A further survival analysis stratified by TME score demonstrated that patients with a high TME score phenotype had a better prognosis than those with a low TME score phenotype (see Figure 1D).
In terms of clinical characteristics, we performed unsupervised hierarchical clustering on 202 survival-related genes, and the significant prognostic effect of the immune subtypes in 160 patients with EC were classified as either high or low TME score subtypes (see Figure 2A and Table S4). The survival difference was independent of grade, gender, purity, ploidy, tumor mutational burden (TMB), and OS status. The DEG analysis revealed 15 differentially expressed miRNAs and 202 differentially expressed mRNAs between the high and low TME score subtypes with a threshold adjusted P<0.05 and a |log2FC| >0.25 for miRNA, and an adjusted P<0.05 and a |logFC| >0.5 for mRNA, respectively.
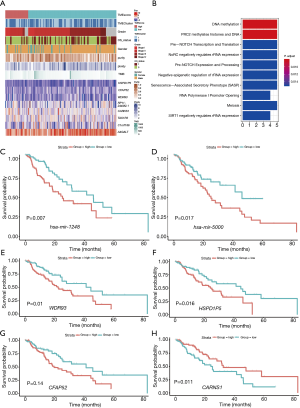
The biological processes of GO terms revealed that differentially expressed mRNAs were mainly enriched in deoxyribonucleic acid (DNA) methylation, polycomb repressive complex 2 (PRC2) methylates histones, and DNA (see Figure 2B). However, we did not find any differentially expressed methylated site. Based on the Cox regression model, 2 differentially expressed miRNAs and 8 differentially expressed mRNAs were identified that had a statistically significant effect on the prognosis of EC. In relation to these, the 2 miRNAs were hsa-mir-1248 and hsa-mir-5000, and the top 4 genes were WDR93, CARNS1, CFAP52, and HSPD1P5. The OS of patients with a high expression level of has-mir-1248 or has-mir-5000 was significantly shorter than that of patients with a lower expression level (see Figure 2C,2D). Similarly, a high expression level of WDR93, CFAP52, and HSPD1P5 was a negative prognostic factor of EC (see Figure 2E-2G), while a high expression level of CARNS1 was a good prognostic factor in EC (see Figure 2H).
Genomic alterations of high and low TME score subtypes of EC
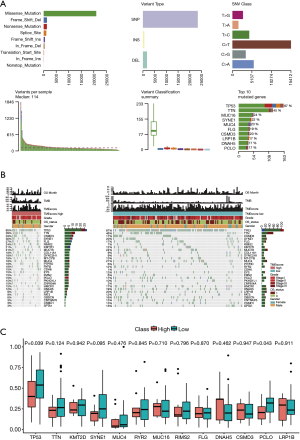
The landscape of the somatic mutations of EC are presented in Figure 3A. Missense mutation mainly caused by SNV was the major type of mutation, and C>T was the major type of base substitution (5). The most frequently mutated genes were TP53 in the high and low TME score subtypes. There were 28 genes with significant differences in mutation frequencies between the high and low TME score subtypes, among which the top 10 differential genes were TP53, TTN, CSMD3, SYNE1, FLG, RIMS2, KMT2D, MUC16, MUC4, RYR2, LRP1B, DNAH5, and PCLO (see Figure 3A, B, C). TTN-AS1, which is a potential diagnostic and prognostic biomarker (25), is highly expressed in ESCC tissues, and promotes ESCC cell proliferation and metastasis (26). PCLO, which is frequently mutated and amplified in ESCC, represents a novel prognostic biomarker and therapeutic target for patients with ESCC (27). These results further confirmed that these high-frequency mutated genes play a key role in the prognosis of EC tumors.
The mutational signature analysis showed that the high TME score subtype was associated with Signature 1, Signature 13, Signature 17, and Signature 29 (see Figure 4A). Conversely, the a TME score subtype was associated with Signature 1, Signature 3, and Signature 17 (see Figure 4B). Signature 1 was associated with the spontaneous deamination of 5-methylcytosine, Signature 3 was associated with DNA- double-strand breaks repair, and Signature 29 was associated with exposure to tobacco mutagens.
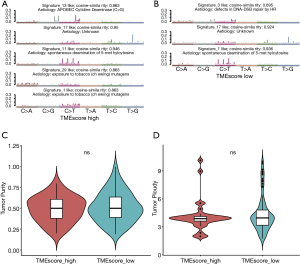
We also investigated genomic instability. The CNVs analysis by GISTIC showed that the significantly amplified regions included 7p, 8q, and 20q. Further, deletions, including at 3p, 5q, and 18q, frequently occurred in the low TME score subtype (see Figure S3A), whereas deletions, including at 9p, 4p, 5q, 18q, and 21q, frequently occurred in the high TME score subtype (see Figure S3B). The minimal common region analysis showed that amplifications of 8q24.21 and 11q13.3, and deletions of 2q22.1, 9p21.3, and 16q23.1 frequently occurred in the high TME score subtype, while amplifications of 11q13.3, and deletions of 7q31.1, 9p21.3, 16q23.1, and 4q22.1 frequently occurred in the low TME score subtype (see Figure S3C and Table S5). Thus, the results indicated a more instable status of chromosomes in patients with the low TME score subtype. Based on the CNV results, the tumor purity and ploidy analysis by ABSOLUTE showed that the estimated tumor purity ranged from 0.19 to 1.00, and tumor ploidy ranged from 1.75 to 10.12 (see Figure 4C,4D and Table S6). Thus, genomic disorder appears to be a common phenomenon in the process of tumorigenesis. There were no significant differences between high and low TME score subtypes in terms of tumor purity and ploidy (Wilcoxon-test, P=0.62 and P=0.55, respectively).
The therapeutic benefits of the TME score
The TME score is a significant biomarker that can add prognostic information beyond that provided by standard clinicopathological properties (6,28). The predictive value of the TME score in EC patients who received adjuvant chemotherapy was examined in a TCGA-ESCA data set. The EC cohort included the mRNA expression levels of patients (n=42) before and after treatment with adjuvant chemotherapy. However, there was no statistically significant differences between patients before and after treatment with adjuvant chemotherapy (P=1.0 for TCGA-ESCA; see Figure S4A). In addition, MSI/dMMR was more frequently detected in well-differentiated tumors and younger patients (≤60 years) (29). We also tried to determine whether the TME score and MSI were correlated. The relationship between the TME score and MSI was identified in EC patients who were examined in the Sensor score and MANTIS (n=157) cohorts. The TME score was not correlated with MSI/dMMR (see Figure S4B,S4C).
The inhibition of the immunological checkpoints that block T-cell inhibitory molecules programmed cell death protein-1 (PD-1) and PD-L1, including pembrolizumab and nivolumab, has emerged as an anti-cancer treatment with unprecedented and synergistic survival benefits (30). Previous research has implied that the Tumor Immune Dysfunction and Exclusion (TIDE) (http://tide.dfci.harvard.edu/) score model was significantly associated with the efficacy of immunotherapy (31). Further, the TIDE model was used to evaluate the clinical efficacy of immunotherapy for high and low TME score subtypes. By Kaplan-Meier analysis, the high TIDE score subtype had much shorter progression free survival then low TIDE score subtype in EC (Figure S4D). However, there was not significant difference in survival prediction efficiency between the low TIDE score subtype and the high TIDE score subtype (Wilcoxon-test, P=0.054) (Figure 5A).
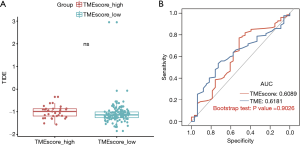
The TMB, which may predict clinical responses to immune checkpoint inhibitors (ICIs) (32), was evaluated by a receiver operating characteristic (ROC) analysis. The TMB is an emerging biomarker for predicting the efficacy of ICIs. The predictive efficiency of the TME score was not significantly different to that of the TMB based on ROC curve analysis (in which the AUC of the TME score was 0.6089 and the AUC of the TMB was 0.6181; P>0.05l; see Figure 5B). Thus, both TME score and TMB can be used to predict the survival of patients with EC, and the results were consistent.
Discussion
Accurate prognostic assessment is critical in selecting the appropriate treatment. Oncologists routinely use the tumor, node, metastasis (TNM) staging system in prognosis. However, as clinical outcomes vary among patients in the same TNM stages, a more accurate prognostic biomarker is needed (15,33,34). Recently, research has revealed that the TME is a promising predictive and prognostic biomarker. IHC has been widely used in various types of cancers, including EC, to characterize the tumor-immune infiltrates within the TME. However, IHC has a number of limitations; for example, it cannot assay cell types simultaneously. In the present study, we applied a bioinformatics method called CIBERSORT that integrates the expression signatures of immune cells to rapidly profile the immune infiltration patterns in EC. Notably, we found a significantly negative correlation between eosinophils and EC survival. Tumor infiltrating eosinophils have been reported in a variety of solid tumors, including EC; however, the function of eosinophils may depend on the cellular composition of the local TME and the presence of a costimulatory factor (35). Eosinophils are regulated by numerous molecules in which interleukin-5 (IL-5) is the most specific and critical cytokine that is responsible for eosinophils biology (36-38). IL-5 has been reported to be positively regulated by miR-1248 (39). MiR-1248 expression in the low TME score group was higher than that of the high TME score group, and high miR-1248 expression was associated with poor survival. These results further support the findings of this study.
We also constructed a classifier, referred to as the TME score, and examined its use as a prognostic tool. We found a significant correlation between the TME score and patients’ survival, which suggests that the TME score is a prognostic biomarker for EC. However, due to the scarcity of RNA-sequencing data sets containing clinical information, validations using other datasets could not be conducted in the current study. Thus, we explored the biological underpinnings of the TME signature to support our findings. Previous studies have suggested that TP53 mutations lead to poor clinical outcomes in a number of cancers (40), and PCLO, which is a frequently mutated and amplified gene in ESCC, contributes to ESCC aggressiveness by stabilizing EGFR and promoting EGFR dependent signaling (27). Our in-depth analysis also revealed a more instable chromosome status in the low TME score group, which may also contribute to the association between low TME scores and poor clinical prognosis.
There are increasing evidences that the TME has substantial promise as a predictive biomarker for the ICI response (41-43). Jiang et al. developed a computational method, called the TIDE tool, to predict the immune checkpoint blockade response (44). However, the lack of treatment information in the study suggests that TIDE tool may not be applicable. Therefore, it is necessary to accept immunotherapy-related RNA data sets to evaluate the value of TME score in predicting the benefit of immunotherapy.
Conclusions
We designed a TME signature to evaluate the comprehensive TME. Our TME score classifier effectively predicted the prognosis of EC patients, which may have important implications for identifying subgroups of EC patients who are at low or high risk of poor survival.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (No. 81871877), the Foundation for Young Scholars of The First Affiliated Hospital of Xiamen University (No. ZLYYA201705) and the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (No. 2020ZX09201005).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-371).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-371). All authors reported that this work had received funding from the National Natural Science Foundation of China (No. 81871877), The Foundation for Young Scholars of The First Affiliated Hospital of Xiamen University (No. ZLYYA201705) and The National Major Scientific and Technological Special Project for “Significant New Drugs Development” (No. 2020ZX09201005), and got technical support from Shanghai Tongshu Biotechnology Co., Ltd. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Ajani JA, D'Amico TA, Bentrem DJ, et al. Esophageal and Esophagogastric Junction Cancers, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2019;17:855-83. [Crossref] [PubMed]
- Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet 2017;390:2383-96. [Crossref] [PubMed]
- Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102. [Crossref] [PubMed]
- Wang F, Liu DB, Zhao Q, et al. The genomic landscape of small cell carcinoma of the esophagus. Cell Res 2018;28:771-4. [Crossref] [PubMed]
- Wang S, Xiong Y, Zhang Q, et al. Clinical significance and immunogenomic landscape analyses of the immune cell signature based prognostic model for patients with breast cancer. Brief Bioinform 2020;bbaa311.
- Zeng D, Li M, Zhou R, et al. Tumor Microenvironment Characterization in Gastric Cancer Identifies Prognostic and Immunotherapeutically Relevant Gene Signatures. Cancer Immunol Res 2019;7:737-50. [Crossref] [PubMed]
- Zheng Y, Chen Z, Han Y, et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun 2020;11:6268. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Fridman WH, Zitvogel L, Sautès-Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717-34. [Crossref] [PubMed]
- Galluzzi L, Buqué A, Kepp O, et al. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell 2015;28:690-714. [Crossref] [PubMed]
- Galluzzi L, Senovilla L, Zitvogel L, et al. The secret ally: immunostimulation by anticancer drugs. Nat Rev Drug Discov 2012;11:215-33. [Crossref] [PubMed]
- Fridman WH, Galon J, Pagès F, et al. Prognostic and predictive impact of intra- and peritumoral immune infiltrates. Cancer Res 2011;71:5601-5. [Crossref] [PubMed]
- Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306. [Crossref] [PubMed]
- Jiang Y, Zhang Q, Hu Y, et al. ImmunoScore Signature: A Prognostic and Predictive Tool in Gastric Cancer. Ann Surg 2018;267:504-13. [Crossref] [PubMed]
- Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909-20. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938-45. [Crossref] [PubMed]
- Stoll G, Bindea G, Mlecnik B, et al. Meta-analysis of organ-specific differences in the structure of the immune infiltrate in major malignancies. Oncotarget 2015;6:11894-909. [Crossref] [PubMed]
- Stoll G, Enot D, Mlecnik B, et al. Immune-related gene signatures predict the outcome of neoadjuvant chemotherapy. Oncoimmunology 2014;3:e27884 [Crossref] [PubMed]
- O’Sullivan JA, Bochner BS. Eosinophils and eosinophil-associated diseases: An update. J Allergy Clin Immunol 2018;141:505-17. [Crossref] [PubMed]
- Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 2010;11:889-96. [Crossref] [PubMed]
- Zheng QX, Wang J, Gu XY, et al. TTN-AS1 as a potential diagnostic and prognostic biomarker for multiple cancers. Biomed Pharmacother 2021;135:111169 [Crossref] [PubMed]
- Lin C, Zhang S, Wang Y, et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin Cancer Res 2018;24:486-98. [Crossref] [PubMed]
- Zhang W, Hong R, Xue L, et al. Piccolo mediates EGFR signaling and acts as a prognostic biomarker in esophageal squamous cell carcinoma. Oncogene 2017;36:3890-902. [Crossref] [PubMed]
- Zhang H, Song J, Dong J, et al. Tumor Microenvironment Analysis Identified Subtypes Associated With the Prognosis and the Tumor Response to Immunotherapy in Bladder Cancer. Front Genet 2021;12:551605 [Crossref] [PubMed]
- Lee HK, Kwon MJ, Ra YJ, et al. Significance of druggable targets (PD-L1, KRAS, BRAF, PIK3CA, MSI, and HPV) on curatively resected esophageal squamous cell carcinoma. Diagn Pathol 2020;15:126. [Crossref] [PubMed]
- Curran MA, Montalvo W, Yagita H, et al. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A 2010;107:4275-80. [Crossref] [PubMed]
- TIDE: Tumor Immune Dysfunction and Exclusion. Available online: http://tide.dfci.harvard.edu/
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Galon J, Pagès F, Marincola FM, et al. Cancer classification using the Immunoscore: a worldwide task force. J Transl Med 2012;10:205. [Crossref] [PubMed]
- Galon J, Pagès F, Marincola FM, et al. The immune score as a new possible approach for the classification of cancer. J Transl Med 2012;10:1. [Crossref] [PubMed]
- Reichman H, Karo-Atar D, Munitz A. Emerging Roles for Eosinophils in the Tumor Microenvironment. Trends Cancer 2016;2:664-75. [Crossref] [PubMed]
- Collins PD, Marleau S, Griffiths-Johnson DA, et al. Cooperation between interleukin-5 and the chemokine eotaxin to induce eosinophil accumulation in vivo. J Exp Med 1995;182:1169-74. [Crossref] [PubMed]
- Rothenberg ME. Eosinophilia. N Engl J Med 1998;338:1592-600. [Crossref] [PubMed]
- Sanderson CJ. Interleukin-5, eosinophils, and disease. Blood 1992;79:3101-9. [Crossref] [PubMed]
- Panganiban RP, Pinkerton MH, Maru SY, et al. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol 2012;1:154-65. [PubMed]
- Wang X, Sun Q. TP53 mutations, expression and interaction networks in human cancers. Oncotarget 2017;8:624-43. [Crossref] [PubMed]
- Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133-50. [Crossref] [PubMed]
- Riaz N, Havel JJ, Makarov V, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017;171:934-949.e16. [Crossref] [PubMed]
- Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568-71. [Crossref] [PubMed]
- Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550-8. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


