
Expression, clinical significance and correlation of RUNX3 and HER2 in colorectal cancer
Introduction
According to the latest epidemiological report, China has an average of 376,000 new patients with colorectal cancer (CRC) every year, about 191,000 people die of CRC, and there is a trend of gradual growth in cases and younger age, with most patients being diagnosed in the middle and late stages, which carries a poor prognosis (1). The pathogenesis of CRC is complex due to its multi-factor and multi-stage co-pathogenesis. RUNT-associated transcription factor 3 (RUNX3) is a tumor suppressor gene that has attracted much attention in CRC research in recent years. Previous studies have shown that its expression is higher in normal tissues than in cancer tissues, especially in digestive tract tumors (2-6). Human epidermal growth factor receptor 2 (HER2) is a well-studied oncogene; its expression in breast cancer and gastric cancer is relatively mature, and its expression is related to prognosis and there is consensus on the effectiveness of it as a therapeutic target. However, there are few studies on HER2 expression in CRC and there are certain differences (Most researchers believe that the high expression of HER2 in CRC is related to the occurrence, development and prognosis of CRC.A few researchers have suggested that HER2 is not associated with the prognosis of CRC) (7-12). This study aimed to jointly detect the expression of RUNX3 and HER2 in CRC to preliminarily explore their correlation and clinical significance. RUNX3 may lead to CRC by negatively regulating HER2 gene-related pathways and detection of RUNX3 and HER2 in CRC could be used as a guide for targeted therapy thus provide experimental basis and scientific basis molecular mechanisms that occur and for targeted therapy of CRC. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-403).
Methods
General data
All CRC specimens surgically resected from the First Affiliated Hospital of Hebei North University from May 2015 to January 2016 were collected, a total of 53 cases, and the adjacent normal tissues >10 cm from the tumor were taken as control samples. Part of each sample was frozen in liquid nitrogen to extract mRNA and proteins. The remaining tissue was fixed with 10% formalin solution for sectioning. There were 28 males and 25 females among the CRC patients; 21 were aged from 41 to 61, 32 were aged from 62 to 84 years; 45 of the patients had high or medium differentiation, 8 patients had low differentiation; there were 44 cases of adenocarcinoma and 9 of mucinous adenocarcinoma; 23 cases were located in the colon and 30 in the rectum; 24 patients were pN+, 29 were pN–; 34 patients were pT I + II, and 19 were pT III + IV. All the diagnoses were confirmed by pathology, and none of the patients underwent chemotherapy, radiotherapy or immunotherapy before surgery. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was reviewed and approved by the Ethics Committee of The First Affiliated Hospital of Hebei North University (No.:K2020291). All patients signed informed consent. Human colorectal adenocarcinoma cell lines RKO and HCT-116 were used for si-RNA interference experiments.
Patient information was collected: sex, age, site of the tumor, tumor tissue type, degree of differentiation, pT stage, pN stage, and Expression levels of RUNX3 and HER2 genes and proteins.
Reagents
For immunohistochemistry, a rabbit SP kit (SP-9001), DAB kit (ZLI-9018, Beijing Zhongshan Jinqiao Biological Co., Ltd.), rabbit-antihuman RUNX3 (EPR20687, Abcam), and rabbit-antihuman HER2 (ER0106, Huaan Bio) were obtained.
For reverse transcription-polymerase chain reaction (RT-qPCR), a total mRNA extraction kit (TR150-50), reverse transcription 5xABScript (RK20402), and SuperReal Fluorescent quantitative kit (2xSYBR Green) (RK21203, Beijing Tianmo Sci & Tech Development Co., Ltd.) were used. Shanghai Sangon Bio designed the synthetic primers.
For western blot, RIPA (Radio Immunoprecipitation Assay), SDS (sodium dodecyl sulfate) gel reagents, SDS-PAGE (SDS polyacrylamide gel electrophoresis), PVDF (polyvinylidene fluoride) membrane, and ECL (electrochemiluminescence) kit were bought from Beyotime Biotechnology. Rabbit-antihuman RUNX3 (AF5188), rabbit-antihuman HER2 (ER0106), and rabbit-antihuman GAPDH (SA30-01) (house-keeping genes) were bought from Huaan Bio. Si-RUNX3, Si-NC and transfection reagents were synthesized and provided by Shanghai Genepharma Company.
Immunohistochemistry
Tissue samples were dehydrated, clarified, paraffin impregnated, and embedded before 4 µm tissue sections were dewaxed, hydrated, antigen repaired, 100 °C for 5 min, 50 °C for 15 min, and cooled at room temperature. Sections were rinsed with phosphate-buffered saline (PBS) for 15 min before dropping reagent each time. The sections were incubated in 0.5% peroxidase at room temperature for 15 min, incubated in goat serum for 30 min at 37 °C, and primary antibody (RUNX3 1 µL + PBS 100 µL + bovine serum albumin (BSA) 3 µL; HER2 1 µL + PBS 100 µL + BSA 3 µL) was added, before placing them in 4 °C refrigerator overnight. On the second day, secondary antibody was added at 37 °C for 30 min, and horseradish peroxidase was added before incubation at room temperature for 15 min. Under the microscope, DAB (1:20) developed color, and when yellow-brown particles were observed, the reaction was terminated. Dyeing with hematoxylin for 3 min, hydrochloric acid and alcohol differentiation was followed by water rinse for 10 min, and the sections were sealed with neutral resin.
The whole section was first observed under low power, then under high power five high-power fields were randomly selected for analysis using ImageJ software. (I) Automated grading of sample staining degree was performed using an IHC Profiler plug-in. According to the average gray value and area percentage (positive cells), 4 scores were evaluated: 3 points (strong positive), 2 points (positive cells), 1 point (weak positive cells), 0 point (negative cells). (II) Counting of positive cells using the Trainable WEKA Segmentation plug-in. According to the percentage of positive cells (accounting for the total number of cells), the number of positive cells <5% was rated as 0, 6–25% as 1, 26–50% as 2, 51–75% as 3, >76% as 4; the product of two scores <3 was rated as negative, and >3 as positive.
RT-qPCR
To investigate the relative expression levels, 5 mg samples of CRC and adjacent normal tissues were taken, 300 µL of lysate were added, the samples were cut into pieces, homogenized, and the supernatant was extracted by centrifugation. Equal volume of anhydrous ethanol was added to the supernatant, centrifuged, 400 µL RNA washing solution was added, and centrifuged again. The supernatant was discarded. Next, 40 µL DNAsel1 reaction solution (DNAsel1 5 µL + DNA digestion solution 35 µL) was added to the precipitate, and incubated at room temperature for 15 min. Pre-wash solution, washing solution and ethanol were added, and then centrifuged and the supernatant was discarded (according to kit instructions). The purity and concentration of total mRNA were determined by UV spectrophotometry after adding 15 µL RNase-free H2O. A260/280 was between 1.8 and 2.1, and the concentration was between 600 and 1,000 ng/µL. For the reverse transcription system, total mRNA 4 µL + 5× ABscript2RT MIX 4 µL + 12 µL RNase-free H2O was processed at 25 °C for 5 min, 42 °C for 15 min, 85 °C for 5 s, and 12 °C for 30 min. GAPDH was used as reference gene for relative quantitative analysis. Primer sequences are shown in Table 1. For the RT-qPCR reaction system: cDNA 2 µL (200 ng) + 2× SYBR Green Fast qPCR Mix 10 µL + 0.4 µL forward primer + 0.4 µL reverse primer + nuclease-free H2O 7.2 µL were used. Reaction conditions were: 1 cycle at 95 °C for 1 min,;5 cycles of 94 °C for 20 s, 55 °C for 30 s, 72 °C for 30 s, and 40 cycles of 94 °C for 15 s, 60 °C for 30 s, Collect the fluorescent. Three samples were taken from each tissue type, and each sample was replicated in three wells. The dissolution curves were all unimodal, without bimodal and with good peak type (peak width <7 grids, peak >800). The obtained CT (cycle threshold) values were usable. The analytical method was 2-ΔΔCt.

Full table
Western blot
For the relative expressions of proteins, 20 mg samples of CRC and adjacent normal tissues were taken, 200 µL RIPA lysate was added, and the samples were shredded, homogenized, and centrifuged. The supernatant was taken, and the total proteins of CRC and adjacent normal tissues were obtained. The BCA (Bicinchoninic Acid Assay) method was used to determine its concentration. Equal volume of buffer was added, the protein was denatured at 100 °C for 5 min, and stored at 4 °C. Preparation of SDS/PAGE separation glue 8% and 10%, concentrated glue 5%. The proteins were separated by electrophoresis and transferred onto the PVDF membrane, which was sealed with TRIS hydrochloric acid buffer (containing 5% BSA) and slowly shaken at room temperature for 2 h. Shaken slowly the PVDF in primary antibody (GAPDH 1:10,000, RUNX3 1:1,000, HER2 1:1,000) was incubated at 4 °C for 12 h, washed with 1× TBST membrane wash solution three times for 10 min each time. The incubation with dropwise add secondary antibody (1:10,000) was slowly shaken at room temperature for 2 h and washed with 1× TBST membrane wash solution three times for 10 min each time. For the ECL method, AI600 was used to obtain the blots, and all samples were repeated for three times. Images were quantitatively analyzed by Image J, and the relative expression of the target protein was represented by the mean value of the target band gray value [IOD (integrated optical density)].
Survival follow-up
Patients were actively followed up by telephone or reexamination at the outpatient department. The last visit was in January 2021. The endpoint was tumor-related death.
In vitro experiments
The RKO cell lines were maintained with Dulbecco modified Eagle medium (DMEM) with the addition of 12% fetal bovine serum (FBS), and the HCT-116 cell lines were maintained with McCoy 5A medium and 12% FBS. The cells were cultured in an incubator at 37 °C with 5% CO2. Trypsin digested the cells in the logarithmic growth phase, the cell density was diluted to 2×105 cells/mL, and the cells were seeded into 6-well plates with 2 mL/well before being incubated overnight. Next, 5 µL si-Runx3, si-NC (sequence shown in Table 2) and transfection reagents were added with 200 µL DMEM and McCoy 5A without FBS, mixed and held for 5 min. The transfection reagents were added into si-RUNX3 and si-NC respectively and mixed, and then stood for 20 min. The medium in the 6-well plate was removed, the plate was washed twice with PBS, 1.5 mL DMEM or McCoy 5A medium without FBS was added to each well, diluent of the mixture was dropped in, and mixed thoroughly. After 6 h of culture, transfection efficiency was observed. The medium containing FBS was changed. After 24 h, lytic solution was added to collect the cells. cDNA and protein was extracted and the relative expression of RUNX3 in the RKO and HCT-116 cells was detected by RT-qPCR and Western blot. HER2 expression was measured by cDNA and protein with knockdown efficiency ≥50% (knockdown efficiency = 1-si-Runx3 relative expression/si-NC relative expression × 100%).

Full table
Statistical analysis
Statistical analysis was performed using SPSS 17.0 statistical software. Measurement data are expressed as mean ± standard deviation, and pairwise comparison was performed by t test. Enumeration data are expressed as percentage, and pairwise comparison was performed by chi-square test. The disease-free survival (DFS) rate was analyzed by Kaplan-Meier, and the difference was analyzed by log-rank. GraphPad 9.0 was used to create graphs, and Pearson analysis was used to analyze the correlation. P<0.05 was considered statistically significant.
Results
Immunohistochemical results
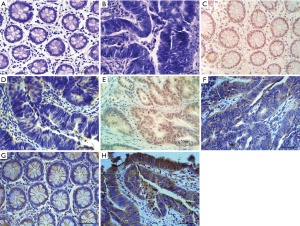
Normal intestinal adipocyte showed uniform size, regular arrangement and clear structure. The adenocarcinoma cells were uneven in size and disordered in arrangement, and the normal tissue structure disappeared (Figure 1A,1B). A total of 18 cases (33.9%) of the 53 cancer tissues were positive for RUNX3, while 43 cases (81.1%) of the normal tissues were positive for RUNX3. The positive expression rate of RUNX3 in CRC was lower than in the adjacent normal tissues, and the difference was statistically significant (P<0.01). RUNX3 was located in the nucleus, and was brown in normal tissue, but negative or dislocation nucleus expressed in CRC (Figure 1C-1E). Among the 53 cases of CRC, 29 (54.7%) were HER2 positive, and in the normal tissue 4 cases (7.5%) were HER2 positive. The positive expression rate of HER2 in CRC was higher than that in the control group, and the difference was statistically significant (P<0.01). HER2 mainly localized in the cell membrane and can be secreted out of the cell. CRC tissues were stained brown, while normal tissues were pale yellow or negatively expressed (Figure 1F-1H).
RT-qPCR results
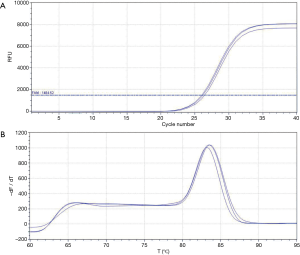
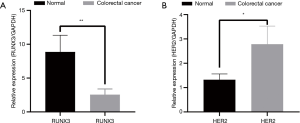
The amplification curve and dissolution curve were good. The dissolution curve was unimodal, the peak width was <7 cells, and the peak height was >1,000 (Figure 2A,2B). The relative expression level of RUNX3 in cancer tissues (2.554±0.8429) was lower than that in adjacent normal tissues (8.864±2.4632), with s statistically significant difference (t=3.481, P=0.005) (Figure 3A). The relative expression level of HER2 in cancer tissues (2.783±0.7444) was higher than that in adjacent normal tissues (1.323±0.2381), with a statistically significant difference (t=−2.656, P=0.017) (Figure 3B).
Western blot results
The relative expression level of RUNX3 protein in the three CRC tissue samples (14,575.667±314.766) was lower than that in the adjacent normal tissues (63,896.667±467.255), with a statistically significant difference (t=131.988, P=0.000). The relative expression level of HER2 protein in cancer tissues (59,142.000±800.989) was higher than that in adjacent normal tissues (4,057.667±648.519), with a statistically significant differences (t=−59.720, P=0.000) (Figure 4).
Relationship between positive expression of RUNX3 and HER2 and clinical features of tumors
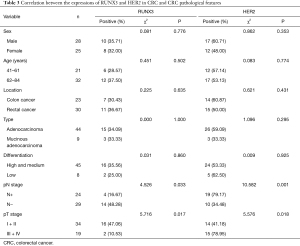
The expressions of RUNX3 and HER2 in CRC did not significantly correlate with sex, age, lesion site, type, etc., but closely correlated with pT and pN staging, as shown in Table 3.
Full table
Survival analysis
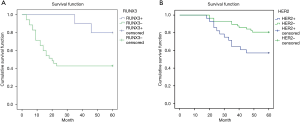
Kaplan-Meier method was used to draw the survival curve of CRC patients, and the survival rate of RUNX3 and HER2 expression was analyzed. The results showed that there were more patients with positive RUNX3 expression in 5-year DFS than those with negative RUNX3 expression (75% vs. 46.15%, P=0.036) (Figure 5A). The patients with HER2 positive expression in DFS at 5 years were fewer than those with HER2 negative expression (53.57% vs. 82.46%, P=0.023) (Figure 5B).
Correlation analysis
The expression of RUNX3 negatively correlated with the expression of HER2 in CRC (r=−0.826, P=0.000) (Figure 6).
In vitro experiments
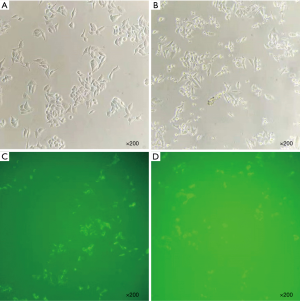
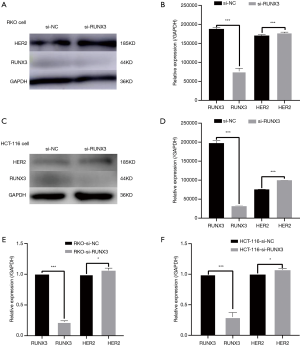
When RUNX3 was knocked down in RKO and HCT-116, HER2 expression increased. The transfection efficiency of si-RUNX3 is shown in Figure 7. The knocking out efficiency of si-RUNX3-1 was 84.7% (P=0.001), that of si-RUNX3-2 was 45.1% (P=0.004), and that of si-RUNX3-3 was 56.1% (P=0.003) (Figure 8). We selected si-RUNX3-1 for subsequent experiments. In RKO and HCT-116 cells knocked down by si-RUNX3, RUNX3 protein expression decreased by 59.3% (P=0.001) and 84.1% (P=0.000),RUNX3 gene expression decreased by 77.4% (P=0.000) and 70.0% (P=0.003), respectively, while HER2 protein expression increased by 3.9% (P=0.000) and 31.7% (P=0.000),HER2 gene expression increased by 6.3% (P=0.041) and 7.2% (P=0.024), respectively (Figure 9).
Discussion
RUNX3 is a tumor suppressor gene that has attracted much attention in recent years. Located on human chromosome 1p36.1, the gene is 67 kb and the protein has a molecular weight of ≈44 kD. RUNX3 has two promoters, P1 and P2, and the P1 promoter contains a large number of T cell binding sites. P2 has cytosine-phosphate-guanine islands and a GC (guanine cytosine) content up to 64%. It is characterized by a GC promoter and is more prone to methylation. RUNX3 protein has 415 amino acid residues, which constitute two subunits (A and B). A binds to the target DNA, and B enhances the binding force of A to maintain the normal regulatory function. The carboxyl terminus is rich in proline and serine, which play an important role in transcriptional regulation (5,10). The human transforming signal transduction molecule Smad protein is the main mediator of the TGF-β(transforming growth factor-β) signal pathway in the cell. Smad’s entry into a specific position must be guided by RUNX3 to jointly activate target genes and regulate cell differentiation, cell cycle, apoptosis and transformation. RUNX3 binds TCF4 and β-catenin in the Wnt pathway, competing for its binding to target DNA and inhibiting carcinogenesis. RUNX3 can regulate the process of epithelial-mesenchymal transformation and claudin-1 protein expression (2,4,13). RUNX3 inhibits gastrointestinal tumor invasion and metastasis by regulating vascular endothelial growth factor (VEGF) (14,15). Highly methylated or point mutated of RUNX3 could reduce RUNX3 expression in CRC (16,17).
HER2, a well-studied proto-oncogene, is the coding product of ERBB2 on chromosome 17 with a molecular weight of ≈185 kD. HER2 protein is a transmembrane glycoprotein with tyrosine kinase activity. HER2 expression is very low in normal human tissues, and high expression usually occurs only during the fetal period. The monomer of HER2 is inactive and the heterodimer has strong growth promoting ability. The upregulation of VEGF by HER2 can promote the formation of new blood vessels and increase the invasiveness of tumors, and the anti-invasion barrier of the body is destroyed (18). The ligand binds to form the dimer, and then phosphorylated phosphokinases are activated. Intracellular adaptor molecules recognize the phosphorylated sites, and intracellular signaling pathways such as Ras/MAPK, PI3K/Akt, or DAG/IP3 are coupled, resulting in cell growth and proliferation. At the same time, a series of negative feedback mechanisms, such as endocytosis, circulation and degradation, dephosphorylation, and serine/threonine phosphorylation in the intimal domain, attenuate the growth signal and destroy the regulatory mechanism, such as HER2 mutation or amplification, leading to continuous growth and carcinogenesis. Tumor proliferation, differentiation and metastasis can also be regulated by STAT, PLCg and other signaling pathways (19).
Previous studies have shown that the expression of RUNX3 in normal tissues is higher than in CRC (2-6). The study of HER2 is relatively mature in breast cancer, and prognosis is related to its expression level. The targeted drug Herceptin has become the standard treatment for patients with HER2-positive breast cancer (20), and its expression in gastric cancer is associated with tumor invasiveness and poor prognosis (21). There are many studies on the expression of RUNX3 and HER2 in CRC, but there are some differences in the reports (7-12). There are few studies of simultaneous investigation of RUNX3 and HER2 expressions in CRC, and the relationship between RUNX3 and HER2 in CRC has not been reported. In this study, immunohistochemistry was used to detect the expression and locations of RUNX3 and HER2. The relative expression level of mRNA was detected at the molecular level by RT-qPCR, and the relative expression level of the proteins was detected by western blot. This study showed that the mRNA and protein of RUNX3 in CRC were lower than in the adjacent normal tissues, while the mRNA and protein of HER2 in CRC were higher than in the adjacent normal tissues.
In this study, survival analysis showed that RUNX3 was associated with 5-year DFS of CRC, and the 5-year DFS of RUNX3-positive CRC patients was higher better than that of -negative CRC patients. The mRNA and protein of RUNX3 were closely related to the diagnosis, treatment and prognosis of CRC. Detection of RUNX3 mRNA and protein can be used in the diagnosis of CRC and can determine prognosis (22). Recently, the detection of serum HER2 has been proved to be a quantifiable examination method by Siravegna et al. (23), indicating that the mRNA relative expression of serum RUNX3 can be an indicator for early screening of CRC.
In this study, the HER2 positive rate in CRC was 54.7%, which was similar to that reported by Wu et al. (24), higher than the 32.4% reported by Sun et al. (7), and lower than the 75% reported by Wen et al. (9), which may be related to whether the samples were fixed in time, whether the samples were repeatedly freeze-thawed (protein degradation results in a negative test), and the judgment of positive results (25). This study proved that HER2 was highly expressed in CRC, and survival analysis showed that the 5-year DFS of CRC was associated with HER2, and that the 5-year DFS of CRC was lower in HER2-positive patients than in negative patients. Detection of HER2 in CRC patients could be used as a guide for targeted therapy, which could benefit them similarly to breast cancer and gastric cancer.
This study revealed a negative correlation between RUNX3 and HER2, and further verified the correlation by cell experiments. The carboxyl terminal of RUNX3 is rich in proline and serine, which play an important role in transcriptional regulation. HER2 protein is a transmembrane glycoprotein with tyrosine kinase activity, which may affect the phosphorylation of RUNX3 proline and serine. This results in the silencing of RUNX3, resulting in the inability of the bound Smad protein to cross the membrane from within the cytoplasm to specific sites. HER2 promotes the generation of new blood vessels by upregulating VEGF and increases tumor invasion. The anti-invasion barrier of the body is destroyed, which may affect RUNX3 ability to regulate the expression of VEGF molecules to inhibit the invasion and metastasis of gastrointestinal tumors. Upregulated Bcl-1, RUNX3 and HER2 are located on the same chromosome. There may be interactions that lead to RUNX3 methylation or point mutation, but the specific interaction mechanism between RUNX3 and HER2 remains to be further studied.
Above all, RUNX3 and HER2 in the diagnosis of CRC and targeted therapy have important significance. In this study, only the gene and protein levels were detected to analyze their expression, correlation and clinical significance of prognosis in CRC tissue. Moreover, the negative regulatory relationship between RUNX3 and HER2 was confirmed by cell experiments. The interaction mechanism is preliminarily discussed. However, this study has not confirmed the specific interaction pathways between the two genes at the extracellular level, nor has it confirmed the relationship between the two genes and the occurrence and development of CRC at the intracellular level (such as tumor formation experiments in nude mice, etc.). This will be a further research direction.
Acknowledgments
Funding: Clinical Medicine Excellent Talents Training Project of Hebei Provincial Finance Department (361009), Medical Science Key Project of Hebei Provincial Science and Technology Department 2015 (152777237), Medical Science Key Project of Hebei Provincial Health and Family Planning Commission (20210519), Mandatory Project of Zhangjiakou Science and Technology Bureau of Hebei Province (1712005D).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-403
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-403
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-403). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of the First Affiliated Hospital of Hebei North University (No.:K2020291) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- National Health Commission of the People's Republic of China. Chinese Protocol of Diagnosis and Treatment of Colorectal Cancer (2020 edition). Zhonghua Wai Ke Za Zhi 2020;58:561-85. [PubMed]
- Ren Y, Cai J, Chen J. The significance of anti-oncogene Runx3 expression in colorectal cancer. Journal of Modern Oncology 2018;1:72-6.
- Xue J, Wu X, Wang L, et al. Expression of RUNX3 in colorectal cancer, adenoma and normal colorectal tissues and its clinical significance. Clin Exp Pathol 2014;30:605-9.
- Wu L, Li Y, Liu H, et al. The role of RUNX3, β-catenin and C-myc in carcinogenesis of colorectal adenoma Chinese Journal of Difficult and Complicated Cases 2018;17:176-179.
- Xue J, Wu XL, Huang XT, et al. Correlation of RUNX3 expression with microvessel density in colorectal adenocarcinoma tissues and clinical significance. Asian Pac J Trop Med 2017;10:98-101. [Crossref] [PubMed]
- Ojima F, Saito Y, Tsuchiya Y, et al. Runx3 transcription factor regulates ovarian functions and ovulation in female mice. J Reprod Dev 2016;62:479-86. [Crossref] [PubMed]
- Sun X, Shen J, Gun-lin H, et al. Relationship between HER2 and Ki-67 expression in colorectal cancer and prognostic factors analysis. J Chongqing Medicine Journal 2018;47:1610-5.
- Chai H, Jin X, Jin X, et al. Microsatellite instability and HER2expression are correlated with the malignancy of colorectal cancer. Basic & Clinical Medicine 2019;39:218-21.
- Wen D, Wang J, Yin Y, et al. The role of research in colorectal cancer and HER2 expression levels, development and invasion and metastasis. Journal of Clinical and Experimental Medicine 2016;15:227-9.
- Richman SD, Southward K, Chambers P, et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J Pathol 2016;238:562-70. [Crossref] [PubMed]
- Cho JH, Lim JY, Cho JY. Survival analysis based on human epidermal growth factor 2 status in stage II-III gastric cancer. World J Gastroenterol 2017;23:7407-14. [Crossref] [PubMed]
- Sawada K, Nakamura Y, Yamanaka T, et al. Prognostic and Predictive Value of HER2 Amplification in Patients With Metastatic Colorectal Cancer. Clin Colorectal Cancer 2018;17:198-205. [Crossref] [PubMed]
- Novellasdemunt L, Foglizzo V, Cuadrado L, et al. USP7 Is a Tumor-Specific WNT Activator for APC-Mutated Colorectal Cancer by Mediating β-Catenin Deubiquitination. Cell Rep 2017;21:612-27. [Crossref] [PubMed]
- Zheng Z, Liu L, Zhan Y, et al. Adipose-derived stem cell-derived microvesicle-released miR-210 promoted proliferation, migration and invasion of endothelial cells by regulating RUNX3. Cell Cycle 2018;17:1026-33. [Crossref] [PubMed]
- Choo SY, Yoon SH, Lee DJ, et al. Runx3 inhibits endothelial progenitor cell differentiation and function via suppression of HIF-1α activity. Int J Oncol 2019;54:1327-36. [Crossref] [PubMed]
- Sun G, Zhang C, Feng M, et al. Methylation analysis of p16, SLIT2, SCARA5, and Runx3 genes in hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e8279 [Crossref] [PubMed]
- Lu DG, Ma YM, Zhu AJ, et al. An early biomarker and potential therapeutic target of RUNX 3 hypermethylation in breast cancer, a system review and meta-analysis. Oncotarget 2017;8:22166-74. [Crossref] [PubMed]
- Gao Z, Song C, Li G, et al. Pyrotinib treatment on HER2-positive gastric cancer cells promotes the released exosomes to enhance endothelial cell progression, which can be counteracted by apatinib. Onco Targets Ther 2019;12:2777-87. [Crossref] [PubMed]
- Wang X, Gu S, Mi L. Structural basis for EGFR transmembrane signaling and its implications in therapeutic development. Science in China 2018;48:735-44.
- Tanner M, Hollmén M, Junttila TT, et al. Amplification of HER-2 in gastric carcinoma: association with Topoisomerase IIalpha gene amplification, intestinal type, poor prognosis and sensitivity to trastuzumab. Ann Oncol 2005;16:273-8. [Crossref] [PubMed]
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [Crossref] [PubMed]
- Zhang L, Yu X, Zheng L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268-72. [Crossref] [PubMed]
- Siravegna G, Sartore-Bianchi A, Nagy RJ, et al. Plasma HER2 (ERBB2) Copy Number Predicts Response to HER2-targeted Therapy in Metastatic Colorectal Cancer. Clin Cancer Res 2019;25:3046-53. [Crossref] [PubMed]
- Wu QB, Sun GP. Expression of COX-2 and HER-2 in colorectal cancer and their correlation. World J Gastroenterol 2015;21:6206-14. [Crossref] [PubMed]
- Shi M, Wang Z, Liu XY, et al. Inactivation of RUNX3 predicts poor prognosis in esophageal squamous cell carcinoma after Ivor-Lewis esophagectomy. Med Oncol 2014;31:309. [Crossref] [PubMed]
(English Language Editor: K. Brown)





