
The predictive value of RNA binding proteins in colon adenocarcinoma
Introduction
Colon cancer is a common malignant tumor of the digestive tract and the fifth leading cause of death from cancer (1). Colon adenocarcinoma (COAD) is the primary pathological type of colon cancer. As a complex heterogeneous disease, the incidence of colon cancer is high and its prognosis is poor. Although treatment of colon cancer has improved with recent advances in medical standards, the 5-year relative survival rate of colon cancer patients is still only 65% (2). Therefore, we need a greater understanding of the molecular mechanism of colon cancer in order to develop more effective diagnostic and treatment methods to improve the prognosis of patients.
RNA binding protein (RBP) is the general term for the protein that binds to RNA, controlling the metabolic process of RNA (3). Its primary role is to mediate RNA maturation, transport, localization, and translation (4,5). RBPs are deregulated in different cancer types, thereby affecting the expression and function of oncoproteins and tumor suppressor proteins (6). It has been reported that RBPs are involved in the expression of a series of genes related to cell growth and proliferation. Changing the expression of these RBPs has been shown to cause physiological defects in cells that lead to cancer development (7). Some specific RBPs are considered to be biological process regulators with versatility and stem cell characteristics, and this feature may lead to tumor heterogeneity. For example, high expression of MSI1 is a sign of poor prognosis in medulloblastoma (8), and overexpression of LIN28B can drive the growth of liver cancer in a mouse model (9). To fully understand the role of RBPs in COAD, we downloaded the clinical and transcriptome data of COAD patients from The Cancer Genome Atlas database (TCGA) database and used a bioinformatics method to find the differentially expressed RBPs in normal and tumor tissues. Based on these RBPs, we built a risk model to predict the prognosis of COAD patients. These differentially expressed RBPs and our model may provide new approaches for diagnosing and treating COAD.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-318).
Methods
Data download and preliminary processing
The RNA sequencing data of 449 COAD samples, 38 normal colon samples, and the corresponding clinical information were downloaded from TCGA database (https://portal.gdc.cancer.gov/). We used Perl software to merge the transcriptome data and clinical information and the limma package (https://www.bioconductor.org/packages/release/bioc/html/limma.html) in R software to extract the RBP expressions from the combined data. Of these, the gene with zero expression in each sample was eliminated. The limma package was also used to identify the differentially expressed RBPs according to the preconditions of |log 2 fold change (FC)| ≥0.5 and P value <0.05. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Enrichment analysis of the selected RBPs
To research the microcosmic mechanisms and functions of the identified RBPs, we firstly conducted a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis on the selected differentially expressed RBPs to find the potential biological pathways. A Gene Ontology (GO) enrichment analysis including the three aspects of biological process (BP), cellular component (CC), and molecular function (MF) was then performed on the RBPs. All enrichment analyses were completed with the Bioconductor package (http://www.bioconductor.org/) in R with a P value <0.05.
Construction and visualization of the protein-protein interaction (PPI) network
The differentially expressed RBPs were entered into STRING software to construct the PPI network and establish the PPI relationship. In addition, Perl was used to obtain the node and network text from the above file. According to these two texts, we used Cytoscape to visualize the PPI network. We obtained PPI subnetworks by using the Molecular Complex Detection (MCODE) program in Cytoscape to analyze the PPI network and then visualize the subnetworks. Finally, GO and KEGG enrichment analyses were performed on the subnetworks, and the enrichment results were considered meaningful when the adjusted P value was less than 0.05.
Establishment and analysis of the prognostic prediction model
We used the createDataPartition function in R software to randomly divide all samples into two groups; an experimental group and a control group. Then we analyzed the differentially expressed RBPs by univariate Cox regression analysis to acquire the candidate genes related to prognosis. We then combined this data with the clinical information and performed a multivariate Cox regression analysis to discover the prognosis-related RBPs. The prognosis prediction model was constructed according to the risk scores of the prognosis-related RBPs in the experimental group. The risk assessment formula in the model was as follows: risk score = coef (coefficient of prognosis-related RBPs) × Exp (expression of prognosis-related RBPs). For each sample, the risk score was the sum of the risk scores of the 10 prognosis-related RBPs. According to the median risk score of the experimental group, samples were divided into high and low-risk groups, and the differences of OS between the two groups were compared. A ROC curve was used to verify the validity of the model. Similarly, we divided the control group into high and low-risk groups according to the median value of the risk score in the experimental group and performed the above steps. The clinical features and risk scores of patients with COAD were analyzed by univariate and multivariate independent prognostic analysis to determine whether the prognostic prediction model could be used as an independent prognostic factor.
Quantification of the COAD prognostic prediction model
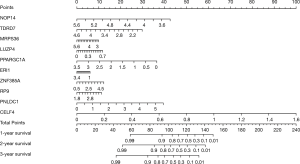
A nomogram was established according to the characteristics of the 10 differentially expressed RBPs. The expression of each RBP corresponded to a score in the nomogram, and the total score of a patient was obtained by adding the scores of the 10 differentially expressed RBPs. The prognostic survival rate of COAD patients was located in the lower part of the nomogram. The nomogram provided a more intuitive understanding of the relationship between the prognosis-related RBPs and the survival of COAD patients.
Further validation of prognosis-related RBPs in terms of survival time and expression level
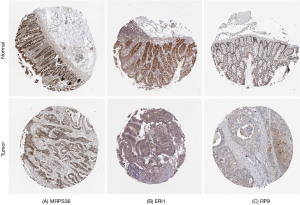
We used the online survival analysis tool, Oncolnc (http://www.oncolnc.org/), to verify the relationship between the expression of the 10 RBPs and the prognosis of COAD. In addition, the immunohistochemical pictures of prognosis-related RBPs in normal and tumor tissues in the Human Protein Atlas (HPA, https://www.proteinatlas.org/) database were used to verify the relationship between the RBPs and COAD prognosis at the translation level.
Statistical analysis
In this study, R 4.0.0 (https://cran.r-project.org/bin/windows/base/old/4.0.0/) was used for statistical analysis of data.The statistical method of wilcoxon rank sum test was used for difference analysis, and the statistical method of Cox survival analysis was used for prognosis correlation analysis.All the above data are based on P<0.05 was statistically significant.
Results
Screening of differentially expressed RBPs in all COAD patients
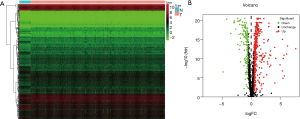
After combining the clinical information and transcriptome data, we obtained a total of 1,493 RBPs (https://cdn.amegroups.cn/static/public/JGO-21-318-1.xlsx). In this study, only 469 differentially expressed RBPs met the inclusion criteria (|log 2 FC| ≥0.5, P<0.05), including 321 high-expression and 148 low-expression RBPs. Figure 1 shows the expression status of these differential genes.
GO and KEGG enrichment analyses of the differentially expressed RBPs
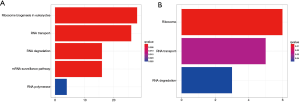
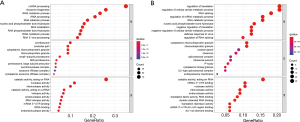
The remaining 469 RBPs were divided into two groups according to their expression levels; a high-expression group and a low-expression group. The KEGG pathway enrichment analysis of the genes in these two groups showed that the RBPs in the high-expression group were mainly concentrated in the ribosome biogenesis in eukaryotes pathway, while the RBPs in the low-expression group were mainly enriched in the ribosome pathway (Figure 2). Results of the GO enrichment analysis are shown in three aspects (Figure 3). In terms of BP, low-expression RBPs were mainly enriched in the regulation of translation. The high-expression RBPs were enriched in ncRNA processing. P-body was the main manifestation of the enrichment for low-expression RBPs in CC, while pre-ribosome was the enrichment form of high-expression RBPs in this aspect. In MF, ribonuclease activity was enriched in upregulated RBPs, and mRNA 3'-UTR was significantly enriched in downregulated RBPs.
Generation and analysis of the PPI network
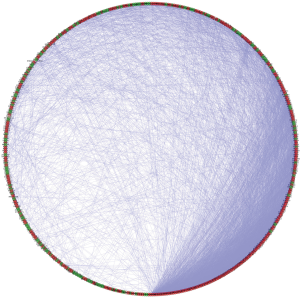
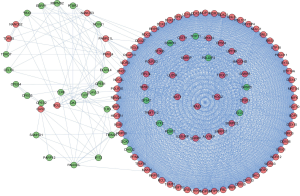
We constructed a PPI network based on the relationship between the differentially expressed RBPs. Figure 4 shows the PPI network after visualization with Cytoscape, containing 442 nodes and 6,631 chains. The MCODE program was used to analyze the PPI network and obtained five subnetworks (Figure 5). Subnetwork 1 contained the largest number of nodes and edges and was therefore the most representative. The RBPs in subnetwork 1 were mainly enriched on the ribosome biogenesis in eukaryotes pathway, and their GO enrichment mainly manifested in ribosome biogenesis, catalytic activity, and acted on RNA and pre-ribosome.
Building and verifying the prognostic model
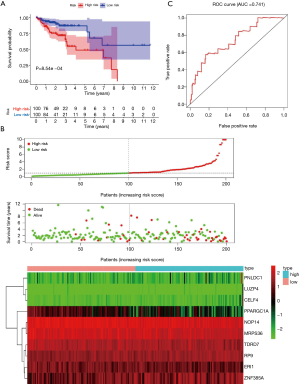
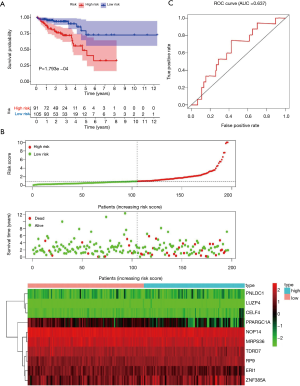
Of the 469 differentially expressed RBPs, 10 prognosis-related RBPs were identified using a univariate Cox regression analysis (Figure 6A, P<0.01). We performed a multiple Cox regression analysis for these 10 alternative RBPs by combining clinical characteristics and survival time and confirmed that all were prognosis-related RBPs in COAD patients (Figure 6B, P<0.001). All samples were divided into two groups; an experimental group and a control group. The construction of the prognostic prediction model was based on the 10 prognosis-related RBPs in the experimental group. We first calculated the risk score of each patient and then divided the patients in the experimental group into high- and low-risk groups according to the median value. The prognosis of patients in the low-risk group was better than that in the high-risk group (Figure 7A,7B, P=8.54e−04). A ROC curve was used to verify the effectiveness of our model. The area under the curve (AUC) was 0.714, which showed that the model worked well (Figure 7C). Using the median risk score of the experimental group, the patients in the control group were also divided into high- and low-risk groups (Figure 8A,8B, P=1.793e−04). The AUC was 0.637, which also verified the accuracy of our model (Figure 8C).

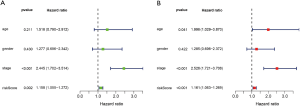
In addition, univariate Cox regression analysis was used to evaluate the relationship between the clinical features of COAD patients, the risk score of the model, and prognosis. The results in Figure 9A show that staging and risk scores are related to the OS of patients. The multivariate Cox regression analysis also showed that age, stage, and risk score were closely related to OS in COAD patients (Figure 9B). These two results confirmed that the model’s risk score could be used as an independent prognostic factor to evaluate the prognosis of COAD patients.
The quantitative model of prognosis in patients with COAD
We constructed a nomogram based on the expression of the 10 prognosis-related RBPs (Figure 10). The scale value in the nomogram was used to calculate the expression of each RBP, and the sum of 10 values represents the score of each patient. We drew a vertical line between the total score and the survival axis to obtain the predicted survival rate of the patient. The nomogram data confirmed that our model was accurate in predicting the prognosis of COAD patients.
Analysis of the effect of RBP expression on the prognosis of COAD
The results from the online survival analysis software showed that a high expression of RP9 was associated with a poorer prognosis for COAD patients, while a high expression of ERI1 and MRPS36 indicated a better prognosis for COAD patients (Figure 11), which is consistent with the HPA results. Compared with the adjacent paracancerous tissues, the staining intensity of RP9 in COAD tissues increased significantly, while the staining intensity of ERI1 and MRPS36 decreased (Figure 12).

Discussion
The incidence and mortality rates of colon cancer have been increasing in recent years, primarily due to changes in diet and environment (10). According to statistics, approximately two-thirds of COAD patients are in an advanced stage of illness at the time of diagnosis, and effective treatment options are limited (11). In recent years, the use of colonoscopy has significantly improved the detection rate of colon cancer, but its invasiveness, potential complications, and cost make it less than ideal for large-scale screening (12). Improvements in colorectal cancer surgery technology and the development of multi-disciplinary treatments, especially chemotherapy using molecularly targeted drugs for unresectable colorectal cancer, will help to improve the survival rate of patients with cancer. Previous studies have shown that malignant tumors, including colon cancer, are determined by genetic changes that affect carcinogenic or tumor-suppressor signaling pathways (13). To diagnose and treat colorectal cancer more effectively, an increasing number of researchers have turned their attention to the microscopic mechanism of tumors. In the field of colorectal cancer, these efforts have promoted the development of precision medicine based on the identification of new biomarkers and novel molecularly targeted drugs to improve prognosis (14). Previous studies have shown that tumor cells can adjust protein expression levels through post-transcriptional mechanisms to adapt to local microenvironments (6). RBPs are involved in the post-transcriptional regulation steps. They affect the structure and interaction of RNA through various mechanisms and play an important role in RNA generation, transportation, and cell localization, thus ensuring the stability of the intracellular environment (15). The imbalance of RBPs affects the occurrence and development of tumors. For example, the epigenetic deletion of CELF2 promotes the growth of breast tumors, leading to a poor prognosis (16). SRSF1 promotes tumor proliferation by enhancing the mRNA stability of DNA ligase 1 in lung cancer (17). The role of RBPs in colon cancer is unclear, so we undertook this study to explore the mechanism of RBPs in colon cancer.
In this study, we performed a KEGG enrichment pathway analysis of 469 differentially expressed RBPs in normal and tumor tissues. We found that RBPs were mainly concentrated in the ribosome biogenesis in eukaryotes pathway and ribosome pathway. It was previously reported that RPL15 was overexpressed in colon cancer through the ribosome biogenesis in eukaryotes pathway and was responsible for synthesizing cell proteins. In addition, the expression of RPL15 was positively correlated with tumor stage. The loss of RPL15 induces apoptosis of colon cancer cells, which may be related to the enlargement of the nucleolus and defects in the internal structure of the nucleolus (dense fiber component or fiber center) caused by RPL15 depletion (18).
Under hypoxic conditions, FGF9 upregulates expression by switching to an internal ribosome entry site (IRES)-dependent translation control, thereby promoting the progression of many cancers, including COAD (19). These reports are consistent with our results, so we hypothesize that these two signaling pathways are part of the mechanism by which RBPs affect COAD prognosis.
The GO functional enrichment analysis of the differentially expressed RBPs indicated that in the upregulated RBPs, the interaction between the Wnt and Hippo pathways may enhance the tumorigenesis potential through a proliferation function mediated by non-coding RNA (RMRP). In addition, the common target genes activated by YAP and β - Catenin, such as RMRP, may provide new tumor targets for tumor therapy (20). In terms of CC, in p53 deficient colon cancer cells, LQ1 has been shown to cause nucleolar stress and impair ribosomal RNA processing, leading to the accumulation of pre-ribosomal RNAs that arrest cells in the G2/M phase and induce early apoptosis (21). In the MF component, cancer cells need to increase the nuclease activity of DNA2 to overcome replication pressure and DNA damage. Studies have proved that DNA2 is overexpressed in a wide range of malignant tumors, including breast cancer, lung cancer, prostate cancer, and colon cancer (22). In this study, the downregulated RBPs were mainly concentrated in the BP of regulation of translation. Previous data has shown that the synthesis rate of general proteins and the expression of several translation components were significantly increased, verifying the potential importance of translation control in tumor progression (23). Phenethyl isothiocyanate (PEITC) has been used to treat human colorectal cancer, and a previous study reported that PEITC increased eukaryotic translation initiation factor 4E (eIF4E) binding protein (4E-BP1) expression and inhibited 4E-BP1 phosphorylation. Translation inhibition is the cause of PEITC-induced apoptosis of cancer cells (24). In terms of CC, the depletion of EXOSC9 reduces the stress resistance and P-body formation of cancer cells. For some types of cancer patients, the increase of EXOSC9 activity is associated with a poor prognosis. Therefore, drugs targeting the RNA exosome complex or EXOSC9 activity may also be useful for these cancers (25). MiR-195-5p is a tumor-suppressor miRNA. It inhibits the expression of the human yes-associated protein 1 (YAP1) by targeting YAP1 mRNA by 3'-UTR, thereby inhibiting the proliferation of colorectal cancer cells (26). In terms of MF, mRNA 3'-UTR binding is the main function of the downregulated RBPs. The results of the GO enrichment analysis showed that the differentially expressed RBPs mainly played a role in the colon through two biological processes, ncRNA processing and regulation of translation. In terms of cell components, pre-ribosome and P-body were the main components of RBPs affecting the prognosis of colon cancer. The mRNA 3'-UTR binding and ribonuclease of RBPs were differentially expressed, indicating that the MF of activity directly impacts the pathological behavior of colon cancer. The above microscopic mechanisms provide a theoretical basis for the prevention and treatment of colon cancer and assist in understanding the occurrence and development of colon cancer so as to develop a more effective diagnosis and treatment protocol.
We also constructed a PPI network of RBPs expressed by the differential markers and obtained a corresponding subnetwork of integration. We found the largest amount of key RBPs in subnetwork 1, and previous studies have found that they were closely related to colon cancer. Liu et al. showed that RNAi silencing of NOB1 can inhibit the growth of colon cancer cells and the G0/G1 cell-cycle arrest, suggesting that NOB1 may be a potential target for the treatment of colon cancer (27). Another study reported that the expression of DHX32 in colorectal cancer was upregulated compared with adjacent normal tissues, and the expression level of DHX32 was related to tumor location, lymph node metastasis, lymph node status, differentiation degree, and Dukes stage (28). Lin et al. showed that DHX32 promoted the proliferation, migration, and invasion of colorectal cancer cells and reduced the sensitivity of 5-FU treatment (29). Similarly, our data also showed that DHX32 upregulated the Wnt pathway and downregulated the expression of pro-apoptotic genes. The results suggest that DHX32 may become a biomarker for the diagnosis and treatment of colorectal cancer. NAT10 promotes the formation of Mn by promoting DNA replication, and NAT10-related Mn activates cGAS-STING-SASP signal transduction and promotes the production of pro-inflammatory factors, thus remodeling the tumor microenvironment and promoting tumor progression. These signals can be enhanced by cell stress and are correlated with the clinicopathological features of colorectal cancer (30). The HIF-1α-VEGF pathway is a classic signal pathway that regulates tumor angiogenesis. Dkc1 directly binds to the promoter region of HIF-1α, promotes the transcription of HIF-1α, increases the expression of HIF-1α and VEGF, and promotes the progress of colorectal cancer (31). Our PPI network module results show that the occurrence and development of COAD are closely related to the ribosome biogenesis in eukaryotes pathway. In terms of BP, the subnetworks are mainly focused on ribosome biogenesis, rRNA metabolic process, ncRNA processing, and ribosomal small subunit biogenesis. RNA helicase activity, snoRNA binding, and 3'-5' RNA helicase activity play important roles in COAD MF, which can be seen in the GO analysis of the PPI network. Therefore, it is likely that these differentially expressed RBPs also affect the development of COAD through some kind of interaction. The study of the RBP interaction process can aid our understanding of the mechanism of colon cancer and assist in finding more effective countermeasures.
Ten prognostic-related RBPs were identified by Cox proportional regression analysis in this study. The risk score of the prognosis-related RBPs in the experimental group was used to establish the prognostic model, which was also applied to the control group. A ROC curve was used to verify the model. We found that the operation of the model in the two groups was meaningful and that our model was effective in predicting the survival of COAD patients. The subsequent nomogram, online survival curve, and immunohistochemical database also verified the validity of our model. Therefore, these prognosis-related RBPs have potential to be translated into clinical applications, but further clinical trials are needed. We know that tumor cells will adapt to changes in the microenvironment by affecting metabolic pathways (32). For instance, one prognostic-related RBP, ARGC1A, is a regulator of some key metabolic pathways and can regulate genes that promote the conversion of glucose to fatty acids to support tumor growth. In colon cancer tissues, it has a medium to high expression (33). Cholesterol acyltransferase 1 (ACAT1) largely promotes the progression of cancer, including cell proliferation, migration and invasion, by participating in abnormal cholesterol metabolism. The corresponding inhibitor, Toll-like receptor 4 (TLR4) can directly targets ACAT1 to inhibit the proliferation and invasion of cancer (34). While the remaining prognostic-related RBPs in colon cancer and their mechanism of action are unclear, it is necessary to continue to explore their role in colon cancer to enrich the prognostic model.
This research has some limitations. Firstly, we used existing information in the TCGA database for analysis and verification and did not verify our findings with in vivo or in vitro experiments; secondly, the construction and verification of the prognostic model was limited to a single TCGA database and therefore lacked cross-validation by information from other databases. Although this study demonstrates that these prognostic RBPs can be used as independent prognostic factors for COAD and are helpful for the identification of new tumor markers and therapeutic targets, these results will require further verification.
Conclusions
In this study, a bioinformatics approach was used to screen out RBPs related to prognosis based on a combination of clinical and transcriptome data, and a prognostic model was constructed. Prognostic-related RBPs appear to be implicated in the occurrence and development of COAD through signaling pathways and MFs. Our model indicates that some specific RBPs are closely related to the prognosis of COAD patients, and their micro-mechanisms may suggest novel approaches for the screening and treatment of colon cancer.
Acknowledgments
Funding: This work was supported by the National Science Foundation of Jiangsu Province of China (BK20191172), the Key Laboratory of Clinical Pharmacy of Jiangsu Province of China (XZSYSKF2020027), the Science and Technology Development Plan of Suzhou City of China (SYS2019007), Project of New Pharmaceutics and Medical Apparatuses of Suzhou City of China (SLJ2021007) and the Gusu Medical Key Talent of Suzhou City of China (GSWS2020005).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-318
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-318
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-318). All authors report that this work was supported by the National Science Foundation of Jiangsu Province of China (BK20191172), the Key Laboratory of Clinical Pharmacy of Jiangsu Province of China (XZSYSKF2020027), the Science and Technology Development Plan of Suzhou City of China (SYS2019007), Project of New Pharmaceutics and Medical Apparatuses of Suzhou City of China (SLJ2021007) and the Gusu Medical Key Talent of Suzhou City of China (GSWS2020005). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Hentze MW, Castello A, Schwarzl T, et al. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 2018;19:327-41. [Crossref] [PubMed]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 2002;3:195-205. [Crossref] [PubMed]
- Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell 2014;54:547-58. [Crossref] [PubMed]
- Pereira B, Billaud M, Almeida R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017;3:506-28. [Crossref] [PubMed]
- Agami R. microRNAs, RNA binding proteins and cancer. Eur J Clin Invest 2010;40:370-4. [Crossref] [PubMed]
- Vo DT, Subramaniam D, Remke M, et al. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am J Pathol 2012;181:1762-72. [Crossref] [PubMed]
- Nguyen LH, Robinton DA, Seligson MT, et al. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 2014;26:248-61. [Crossref] [PubMed]
- Yang C, Zhang Y, Xu X, et al. Molecular subtypes based on DNA methylation predict prognosis in colon adenocarcinoma patients. Aging (Albany NY) 2019;11:11880-92. [Crossref] [PubMed]
- Corvino A, Corvino F, Radice L, et al. Synchronous mucinous colonic adenocarcinoma and multiple small intestinal adenocarcinomas: report of a case and review of literature. Clin Imaging 2015;39:538-42. [Crossref] [PubMed]
- Ahmed FE. miRNA as markers for the diagnostic screening of colon cancer. Expert Rev Anticancer Ther 2014;14:463-85. [Crossref] [PubMed]
- Liu W, Li H, Hong SH, et al. Olfactomedin 4 deletion induces colon adenocarcinoma in ApcMin/+ mice. Oncogene 2016;35:5237-47. [Crossref] [PubMed]
- Nagasu S, Sudo T, Kinugasa T, et al. Y-box-binding protein 1 inhibits apoptosis and upregulates EGFR in colon cancer. Oncol Rep 2019;41:2889-96. [Crossref] [PubMed]
- Glisovic T, Bachorik JL, Yong J, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008;582:1977-86. [Crossref] [PubMed]
- Piqué L, Martinez de Paz A, Piñeyro D, et al. Epigenetic inactivation of the splicing RNA-binding protein CELF2 in human breast cancer. Oncogene 2019;38:7106-12. [Crossref] [PubMed]
- Martínez-Terroba E, Ezponda T, Bértolo C, et al. The oncogenic RNA-binding protein SRSF1 regulates LIG1 in non-small cell lung cancer. Lab Invest 2018;98:1562-74. [Crossref] [PubMed]
- Dong Z, Jiang H, Liang S, et al. Ribosomal Protein L15 is involved in Colon Carcinogenesis. Int J Med Sci 2019;16:1132-41. [Crossref] [PubMed]
- Chen TM, Shih YH, Tseng JT, et al. Overexpression of FGF9 in colon cancer cells is mediated by hypoxia-induced translational activation. Nucleic Acids Res 2014;42:2932-44. [Crossref] [PubMed]
- Park J, Jeong S. Wnt activated β-catenin and YAP proteins enhance the expression of non-coding RNA component of RNase MRP in colon cancer cells. Oncotarget 2015;6:34658-68. [Crossref] [PubMed]
- Pecoraro A, Virgilio A, Esposito V, et al. uL3 Mediated Nucleolar Stress Pathway as a New Mechanism of Action of Antiproliferative G-quadruplex TBA Derivatives in Colon Cancer Cells. Biomolecules 2020;10:583. [Crossref] [PubMed]
- Kumar S, Peng X, Daley J, et al. Inhibition of DNA2 nuclease as a therapeutic strategy targeting replication stress in cancer cells. Oncogenesis 2017;6:e319 [Crossref] [PubMed]
- Holland EC, Sonenberg N, Pandolfi PP, et al. Signaling control of mRNA translation in cancer pathogenesis. Oncogene 2004;23:3138-44. [Crossref] [PubMed]
- Hu J, Straub J, Xiao D, et al. Phenethyl isothiocyanate, a cancer chemopreventive constituent of cruciferous vegetables, inhibits cap-dependent translation by regulating the level and phosphorylation of 4E-BP1. Cancer Res 2007;67:3569-73. [Crossref] [PubMed]
- Yoshino S, Matsui Y, Fukui Y, et al. EXOSC9 depletion attenuates P-body formation, stress resistance, and tumorigenicity of cancer cells. Sci Rep 2020;10:9275. [Crossref] [PubMed]
- Sun M, Song H, Wang S, et al. Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J Hematol Oncol 2017;10:79. [Crossref] [PubMed]
- Liu Y, Huang H, Yuan B, et al. Lentivirus-mediated knockdown of NOB1 suppresses the proliferation of colon cancer cells. Z Gastroenterol 2014;52:429-35. [Crossref] [PubMed]
- Huang C, Liang X, Huang R, et al. Up-regulation and clinical relevance of novel helicase homologue DHX32 in colorectal cancer. J Exp Clin Cancer Res 2009;28:11. [Crossref] [PubMed]
- Lin H, Liu W, Fang Z, et al. Overexpression of DHX32 contributes to the growth and metastasis of colorectal cancer. Sci Rep 2015;5:9247. [Crossref] [PubMed]
- Cao Y, Yao M, Wu Y, et al. N-Acetyltransferase 10 Promotes Micronuclei Formation to Activate the Senescence-Associated Secretory Phenotype Machinery in Colorectal Cancer Cells. Transl Oncol 2020;13:100783 [Crossref] [PubMed]
- Hou P, Shi P, Jiang T, et al. DKC1 enhances angiogenesis by promoting HIF-1α transcription and facilitates metastasis in colorectal cancer. Br J Cancer 2020;122:668-79. [Crossref] [PubMed]
- Shlomi T, Benyamini T, Gottlieb E, et al. Genome-scale metabolic modeling elucidates the role of proliferative adaptation in causing the Warburg effect. PLoS Comput Biol 2011;7:e1002018 [Crossref] [PubMed]
- Alix-Panabières C, Cayrefourcq L, Mazard T, et al. Molecular Portrait of Metastasis-Competent Circulating Tumor Cells in Colon Cancer Reveals the Crucial Role of Genes Regulating Energy Metabolism and DNA Repair. Clin Chem 2017;63:700-13. [Crossref] [PubMed]
- Ye K, Wu Y, Sun Y, et al. TLR4 siRNA inhibits proliferation and invasion in colorectal cancer cells by downregulating ACAT1 expression. Life Sci 2016;155:133-9. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


