
Helicobacter pylori-induced protein tyrosine phosphatase receptor type C as a prognostic biomarker for gastric cancer
Introduction
Helicobacter pylori (H. pylori) has been reported to have a close association with some digestive system diseases, such as chronic gastritis, stomach ulcer, gastric cancer, and pathological gastric mucosa-related lymphoid tissue lymphoma (1-3). As a Gram-negative bacteria, H. pylori could stably grow in a strong acid environment, and some toxins could be induced by H. pylori, including cyclooxygenase-2, cytotoxin-associated gene A (CagA), urease, and inducible nitric oxide synthase (4-7). These toxins subsequently contribute to the gastric mucosa damage. The long-term colonization of H. pylori in the stomach could also disrupt immune imbalance and induce inflammatory responses (8,9). Some pro-inflammatory factors can destroy the gastric mucosal barrier and promote the occurrence and development of gastrointestinal diseases (10,11).
Previous studies have shown the close correlation between H. pylori-induced immune response and the occurrence of gastric cancer, especially for CagA-positive and vacuolating toxin A (VacA)-positive H. pylori infection (12,13). However, the in-depth molecular mechanism underlying the transition from inflammation to gastric cancer is still unclear. The aim of the present study was to identify the key regulator in H. pylori-related gastric cancer and to study the expression level and clinical value of the indicated key regulator in gastric cancer. In brief, we first identified key regulators in the occurrence of H. pylori infection, and the bioinformatics analysis identified the protein tyrosine phosphatase receptor type C (PTPRC), a member of the protein tyrosine phosphatase family, which regulates a number of biological activities, including proliferation, differentiation, and mitosis (14). Previously published literature has also shown that PTPRC, as a kind of protein tyrosine phosphatase (PTP) family, could regulate the T- and B-cell antigen receptor signaling (15,16). As a hub gene, PTPRC is involved in the biological regulation of H. pylori infection and has a close association with gastric cancer. In the present study, we demonstrated that the PTPRC expression level could be induced by H. pylori infection in gastric mucosa tissues. The overexpression of PTPRC in gastric cancer was also validated. We also evaluated the potential application of PTPRC as a prognostic biomarker for gastric cancer. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-305).
Methods
Reagents
H. pylori strain 26695 and Gastric adenocarcinoma AGS cells and BGC-823 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Dulbecco’s modified Eagle’s medium, fetal bovine serum, and penicillin-streptomycin were purchased from Thermo Scientific (Waltham, MA, USA). The total RNA isolation kit, first-strand cDNA synthesis kit, and SYBR quantitative reverse transcription polymerase chain reaction (qRT-PCR) kit were obtained from Yeasen Biotechnology (Shanghai, China). The primers were synthesized by Invitrogen (Carlsbad, CA, USA). The storage of materials was performed according to the manufacturers’ instructions.
Gene Expression Omnibus (GEO) analysis
Microarray data were obtained from the GEO database. The GSE6143 dataset containing RNA sequence data of gastric mucosa tissues with or without H. pylori infection was used to identify differentially expressed genes (DEGs) in gastric mucosa tissues (17). The statistical threshold was set at P<0.05 and >2 fold change. The volcano plot displayed the top 10 DEGs. The GSE13911 dataset was used to evaluate the PTPRC expression level in gastric mucosa tissues and gastric tumor tissues (18).
Gene expression using the Oncomine database
The mRNA levels of the indicated genes in Pan-Cancer was conducted using Oncomine, according to previous reports (19,20). P<0.05 and >2 fold change were used as the threshold of statistical difference. The gene rank was higher than the top levels of 10%. “DErrico gastric” referred to the significant expression difference in gastric cancer, and the reporter probe was 1569830_at, which was also included in the GSE13911 dataset (18).
Enrichment analysis using bioinformatics
Enrichment analysis of DEGs was done through the Metascape web-based portal using the Kyoto Encyclopedia of Genes and Genomes pathway (KEGG), Gene Ontology (GO) biological processes, Reactome gene sets, and canonical pathways (21). The protein-protein interaction was analyzed using Metascape and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; https://string-db.org/) (22). The co-expression network was made.
The Cancer Genome Atlas (TCGA) analysis
The transcriptome data of TCGA with gastric cancer was conducted using the UALCAN portal (http://ualcan.path.uab.edu/) and Gene Expression Profiling Interactive Analysis (GEPIA) tool (http://gepia2.cancer-pku.cn/) (23,24). A total of 408 cases of gastric tumor tissues were included to test the PTPRC expression compared with normal gastric tissues, including the adjacent gastric tissues and Gene Tissue Expression (GTEx) gastric tissues (25). The PTPRC expression at different disease stages was achieved by GEPIA, and other pathological features, such as H. pylori infection, tumor grade, lymph node metastasis, and histological types, were also included to analyze the expression levels of PTPRC. The study was approved by ethics board of Cancer Hospital Affiliated to Guangzhou Medical University. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Individual consent for this retrospective analysis was waived.
qRT-PCR assay
The mRNA levels of AGS and BGC-823 cells were tested by qRT-PCR assay. The cells were treated with H. pylori strain 26695 for 4 h, and the cells were collected and lysed with total RNA isolation kit according to the manufacturer’ guidelines. The total RNA was washed with 75% ethanol and dissolved in H2O without RNase. The cDNA was synthesized by the first-strand cDNA synthesis kit. qRT-PCR was conducted using the SYBR Green kit (supplied with high Rox). The 2-ΔΔCT method was used to analyze the relative gene expression levels. All in vitro experiments were performed in triplicate by the first author.
The detailed sequence of primers was as follows: Tumor necrosis factor receptor superfamily member 7 (TNFRSF7) (left primer: 5'-CAGAGAGGCACTACTGGGCT-3', right primer: 5'-CGGTATGCAAGGATCACACT-3'), lactotransferrin (LTF) (left primer: 5'-CCCAGGAACCGTACTTCAGC-3', right primer: 5'-GTGCCACAACGGCATGAGA-3'), GM-CSF receptor (CSF2RB) (left primer: 5'-CTCCTTTGGCCTATTCTACAAGC-3', right primer: 5'-TGAACAGAGACGATGTATTGGC-3'), PTPRC (left primer: 5'-TTGAGCGACAGGAGGATGAG-3', right primer: 5'-GACGCCTCTCCACATTGCT-3'), and Epithelial membrane protein 3 (EMP3; left primer: 5'-GGAGGTCTCTTCTATGCCACC-3', right primer: 5'-AGGATCTCCTCGGCGTGAAT-3').
Survival analysis using the Kaplan-Meier plotter
The Kaplan-Meier plotter (http://kmplot.com/analysis/) was used for the survival analysis (26). Gastric cancer patients were divided into the high expression group and low expression group by the best cut-off. The log-rank statistical method was used to determine the statistical difference. Cox proportional hazard analysis was also expressed with hazard ratio (HR).
Statistical analysis
All data were expressed as mean ± standard deviation. The difference between two groups was calculated using unpaired Student’s t-test. The correlation analysis between two factors was achieved by the Pearson method, and R>0.5 was considered the significant positive correlation. Statistical differences between multiple groups were determined using F-test with Pr analysis; Pr is the P value associated with the F statistic of a given effect and test statistic. The survival analysis was performed by the Kaplan-Meier method with log-rank test, and the HR was adjusted to the survival analysis. P<0.05 was considered as a statistically significant difference. Data processing was conducted with GraphPad Prism 8.0 (La Jolla, CA, USA).
Results
Identification of significant genes associated with H. pylori infection in gastric mucosa
A total of 24 cases of gastric mucosa tissues were analyzed to determine the effect of H. pylori infection, including 8 cases of H. pylori-negative and 16 cases of H. pylori-positive gastric mucosa tissues. The significant DEGs are shown in Figure 1A. To determine significant DEGs associated with H. pylori infection, a volcano plot was created (Figure 1B), and the top 5 increased or decreased genes were as follows: TNFRSF7, LTF, CSF2RB, PTPRC, EMP3, MAPK7, insulin-like growth factor binding protein 2 (IGFBP2), angiopoietin (ANG), platelet-derived growth factor subunit A (PDGFA), and DTR. To fully understand the expression pattern of that these DEGs, the PaGenBase database was used for the enrichment analysis. The results showed these DEGs were mainly expressed in the spleen, blood, bone marrow, thymus, placenta, colorectal adenocarcinoma, smooth muscle, and lung tissues. Cell-specific analysis showed that these DEGs were also highly expressed in lymphoma Burkitt’s Raji, Roswell Park Memorial Institute (RPMI) 8226, bronchial epithelial cells, adipocyte, breast cell, human umbilical vein endothelial cells (HUVEC), CD56-positive natural killer cells and human acute lymphoblastic leukaemia Cells MOLT4 (Figure 1C). As the most important organism tissues regulating the biological process of immune response, the spleen, blood, bone marrow, and thymus were also enriched in the H. pylori-infected gastric mucosa, indicating potential cross-talk between microflora and immune response in the regulation homeostasis of the gastrointestinal tract.

Immune-related pathway enrichment in gastric mucosa with H. pylori infection
As shown in Figure 1, DEGs were identified in gastric mucosa tissues with H. pylori-negative and -positive infection, and the expression pattern enrichment analysis showed that these DEGs have potential regulation in the process of immune response. To further understand the enrichment network of these DEGs in the process of H. pylori infection, the DEGs underwent process enrichment analysis using KEGG pathway, GO biological processes, Reactome gene sets, and canonical pathways, and the top 10 enrichment networks showed that these H. pylori-induced DEGs were significantly associated with cytokine-cytokine receptor interaction, signaling by interleukins, leukocyte cell–cell adhesion, peptidyl-tyrosine phosphorylation, leukocyte migration, apoptotic signaling pathway, phosphoinositide 3-kinase (PI3K)-Akt signaling pathway, Nuclear factor ĸB (NF-ĸB) signaling pathway, and cancer and leishmaniasis pathways. These findings indicated that exposure of H. pylori could induce immune stress response. These significant DEGs were also enriched in the network of cancer pathways, PI3K-Akt signaling pathway, NF-ĸB signaling pathway, positive regulation of cell migration, cell adhesion molecules, and angiogenesis, which are considered common pro-oncogenic biological mechanisms (Figure 2). These findings indicated that H. pylori infection is an important mediator in gastric cancer tumorigenesis. Considering the enriched immune-related pathways and pro-tumorigenesis probability by H. pylori, the potential underlying mechanism between inflammation and cancer transition was also analyzed. The protein-protein interaction analysis was done to identify the core molecular components in the enrichment network of DEGs induced by H. pylori. The results indicated 9 enrichment components, including the toll-like receptor (TLR) signaling pathway, RAF/MAP kinase cascade, positive regulation of leukocyte cell-cell adhesion, signaling by receptor tyrosine kinases, response to tumor necrosis factor, focal adhesion, PS1 pathway, T-cell receptor signaling pathway, and cellular responses to stress, which were generally consistent with the GO analysis (Figure 3).
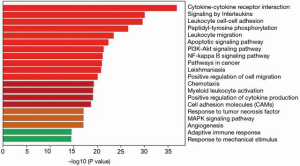
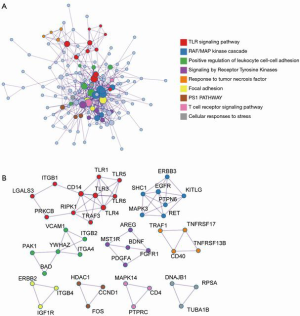
Identification of significant DEGs in gastric mucosa with H. pylori infection
As mentioned earlier, infection is an essential factor that disrupts gastrointestinal homeostasis, and the significant DEGs were identified in Figure 1. We directly display the transcriptional expression level of core genes during the H. pylori infection. The top 5 increased genes are shown in Figure 4A,B,C,D,E and are as follows: TNFRSF7, LTF, CSF2RB, PTPRC, and EMP3. The most significant downregulated genes were MAPK7, IGFBP2, ANG, PDGFA, and DTR (Figure 4F,G,H,I,J). The mRNA expression of these genes in gastric mucosa tissues was significantly decreased with H. pylori infection. As shown in previously published studies (27), TNFRSF7, LTF, CSF2RB, PTPRC, and DTR are closely involved in the regulation of immune function. Moreover, EMP3, IGFBP2, ANG, and PDGFA are common regulators in cell-cell interaction, including cell proliferation, migration, and adhesion. All of these results were consistent with the enrichment analysis results.
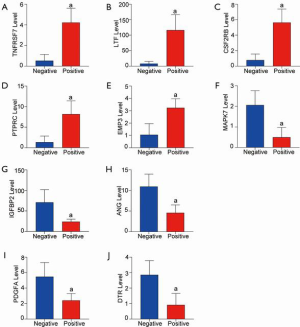
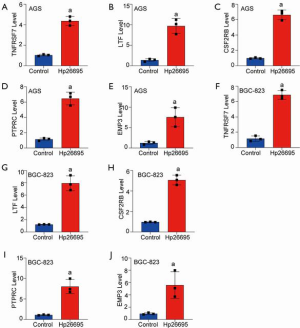
Validation of DEGs in the H. pylori strain-induced cell model
As shown in Figures 1 and 4, TNFRSF7, LTF, CSF2RB, PTPRC, and EMP3 expression in gastric mucosa tissues was significantly increased when infected with H. pylori. The significant upregulated genes might play an essential role in H. pylori cellular response. Therefore, the upregulated regulators were included in the present study, and to further confirm the results of RNA sequence results, AGS and BGC-823 cells were treated with H. pylori strain 26695. The results revealed that after the exposure of H. pylori infection for 4 h in AGS and BGC-823 cell lines, TNFRSF7, LTF, CSF2RB, PTPRC and EMP3 mRNA levels were notably increased compared with the negative control, which were consistent with the results of RNA sequence with H. pylori infection in normal gastric mucosa tissues (Figure 5).
Overexpression of PTPRC in gastric cancer
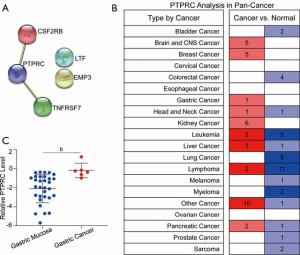
The former RNA sequence results and H. pylori-infected AGS and BGC-823 in vitro cell model both demonstrated that TNFRSF7, LTF, CSF2RB, PTPRC, and EMP3 expression could be increased significantly by H. pylori infection. To further identify the core regulators in these 5 genes, the protein-protein interaction was confirmed in STRING database, and the results indicated that PTPRC was co-expressed with CSF2RB and TNFRSF7, suggesting that PTPRC was the core regulator in these 5 genes during H. pylori infection (Figure 6A). Considering the importance of H. pylori infection in tumorigenesis, especially in the gastric cancer, and to fully understand the role of PTPRC in the development of cancer, the PTPRC analysis in Pan-Cancer was conducted in the Oncomine database. As shown in Figure 6B, PTPRC was overexpressed in brain and central nervous system cancer, breast cancer, gastric cancer, kidney cancer, and other tumor types. As demonstrated earlier, PTPRC is a H. pylori-related mediator and an important risk factor for gastric cancer; therefore, the expression and clinical significance of PTPRC in gastric cancer has also been included in our subsequent study. To directly confirm the difference of PTPRC expression in gastric cancer, the GSE13911 dataset was included, and the data revealed that PTPRC expression increased significantly in gastric cancer tissues compared with normal gastric mucosa tissues (P<0.01) (Figure 6C). Based on these results, H. pylori-induced PTPRC is highly expressed in gastric cancer and might be an important mediator in the regulation of H. pylori-related gastric cancer.
PTPRC expression is positively correlated with CSF2RB and TNFRSF7 in gastric cancer
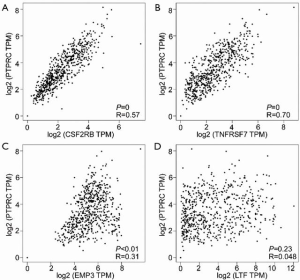
The data showed that PTPRC expression was significantly increased in H. pylori-infected gastric mucosa, and the protein–protein interaction network analysis showed PTPRC and CSF2RB or TNFRSF7 were co-expressed, suggesting the core role of PTPRC in the process of H. pylori infection. Furthermore, PTPRC was overexpressed in gastric cancer. Consequently, the correlation analysis between PTPRC and other significant DEGs was conducted in gastric cancer to interpret the core role of PTPRC in gastric cancer. As the results showed, PTPRC expression was significantly positively correlated with CSF2RB (R=0.57, P<0.001) (Figure 7A) and TNFRSF7 (R=0.70, P<0.001) (Figure 7B). PTPRC had a correlation with EMP3 (R=0.31, P<0.01) (Figure 7C). However, there was no significant correlation between PTPRC and LTF (R=0.048, P=0.23) (Figure 7D). These data were consistent with the results of the bioinformatic analysis using STRING, and PTPRC was further confirmed as a core gene in H. pylori infection and the oncogenesis of gastric cancer.
Overexpression of PTPRC is closely associated with the development of gastric cancer
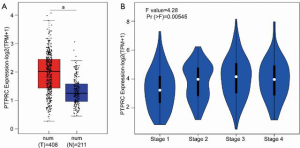
PTPRC was identified as a H. pylori-induced mediator, which was preliminarily studied in the GSE13911 gastric cancer dataset. To validate the result about overexpression of PTPRC in gastric cancer, the much more samples about TCGA gastric cancer database was included in Figure 8A, PTPRC was identified to be overexpressed in gastric cancer tissues than normal tissues, including adjacent normal gastric tissues and normal gastric mucosa tissues from GTEx database. Therefore, our hypothesis was that the overexpression of PTPRC in gastric cancer might be closely associated with the development of gastric cancer. To demonstrate the potential role of PTPRC in the development of gastric cancer, the expression of PTPRC in gastric tumor tissues at different disease stages was analyzed (Figure 8B). The data indicated that PTPRC expression was closely associated with disease stage and PTPRC expression increased with the disease stage in gastric cancer, indicating that PTPRC might be an important factor in the development of gastric cancer.
PTPRC is closely associated with the pathological factors of gastric cancer
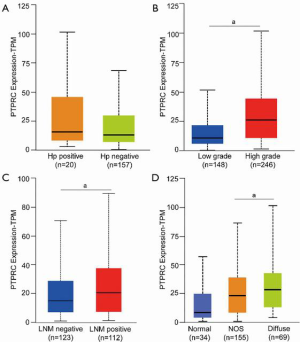
To fully understand the role of PTPRC in gastric cancer, the association between PTPRC expression with some pathological factors was analyzed. The data indicated that H. pylori infection could induce PTPRC expression in gastric mucosa tissues, and the overexpression of PTPRC in gastric cancer was also confirmed. Considering the important role of H. pylori in the oncogenesis and development of gastric cancer, the PTPRC expression in gastric tumor tissues with or without H. pylori infection was confirmed, and the data revealed that PTPRC expression was slightly higher in H. pylori-positive gastric tumor tissues than in H. pylori-negative tumor tissues (Figure 9A). The association between tumor grade and PTPRC levels is shown in Figure 9B, where PTPRC was overexpressed in high-grade gastric tumor tissues than low-grade tissues (P<0.001) (Figure 9B). Lymph node metastasis was included in the subsequent study, in which PTPRC expression was found to increase in gastric tumor tissues with lymph node metastasis compared with those tumor tissue without lymph node metastasis (P<0.001) (Figure 9C). Histological type was also an important factor that was found to contribute to the different prognosis performance of gastric cancer patients (28). As shown in Figure 9D, PTPRC level was higher in diffuse-type gastric tumor tissues than those tissues with not otherwise specified histological types (P<0.001). These findings indicated that PTPRC expression was closely associated with the progression of gastric cancer, especially for those patients with diffuse histological type.
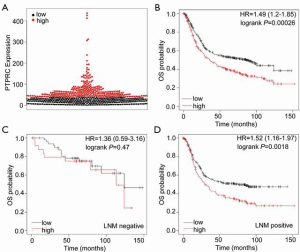
High expression of PTPRC is a poor prognostic factor for gastric cancer
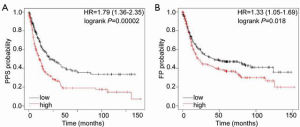
The overexpression of PTPRC in gastric cancer showed a close association with the progression of gastric cancer, such as H. pylori infection, disease stage, tumor grade, lymph node metastasis, and histological type. Therefore, we hypothesized that PTPRC might act as a poor prognostic biomarker for gastric cancer. To confirm this hypothesis, TCGA gastric cancer database was used for the survival analysis. First, the total number of patients with gastric cancer as divided into the high PTPRC expression group and low PTPRC expression group according to PTPRC expression level, and the best cut-off value was used as the threshold for the subgroup (Figure 10A). The overall survival analysis is shown in Figure 10B. Gastric cancer patients with high PTPRC expression had a lower survival rate (log-rank P=0.00026), and the Cox proportional hazard analysis showed that higher PTPRC expression was a risk factor for gastric cancer patients (HR =1.49, 95% confidence interval: 1.2–1.85). Considering the close association between PTPRC levels and gastric cancer progression, an overall analysis of gastric cancer patients with or without lymph node metastasis was conducted. The results demonstrated that PTPRC was a poor prognostic factor for gastric cancer patients with lymph node metastasis (Figure 10C). However, in those gastric cancer patients with no lymph node metastasis, there was no significant difference in the overall survival rate of high or low PTPRC (Figure 10D). These results suggest that the overexpression of PTPRC could act as a poor prognostic biomarker for gastric cancer patients. However, the prognostic evaluation of patients at advanced stage is still difficult; therefore, the post-progression survival (PPS) and first progression (FP) survival analyses were conducted. As shown in Figure 11, the survival rate of gastric cancer patients with high PTPRC expression was significantly lower than that of patients with lower PTPRC expression in both the PPS and FP survival analyses. Therefore, the PTPRC-based prognostic analysis could be a reminder during the treatment of gastric cancer. It is favorable for the improvement of life, especially for gastric cancer patients at advanced stage.
Discussion
H. pylori infection is a lifelong disease that contributes to the chronic gastritis, gastric ulcer, and even gastric cancer. Current studies show the mechanism of the persistence of H. pylori infection that chronic H. pylori infection could induce a Th1-based immune response in gastric mucosa, and then the function of T helper (Th) 2 could be inhibited, thus the local B cells in the gastric mucosa cannot secrete the enough immunoglobulin A to clear the H. pylori. However, the detailed mechanism of H. pylori infection in the cross-talk between gastritis and gastric cancer is still unclear. Therefore, to identify the potential key mediators in the regulation from inflammation to tumorigenesis is important to understand the pathogenic mechanism of H. pylori.
In the present study, we identified DEGs in gastric mucosa with and without H. pylori infection (Figure 1). The bioinformatics analysis showed these DEGs were enriched in immune-related pathways and some cancer pathways (Figures 2,3). As significant DEGs might contribute to the progression of gastric cancer, the most significantly increased genes were also identified and used in an in vivo cell experiment model with AGS and BGC-823 cell lines (Figures 4,5). The results confirmed that TNFRSF7, LTF, CSF2RB, PTPRC, and EMP3 could be induced by H. pylori infection. As shown in previous studies, TNFRSF7, a member of tumor necrosis factor-receptor superfamily, is involved in the regulation of T-cell immunity (27). LTF is an important component of non-specific immune system with antimicrobial activity (28). CSF2RB is a paralog of interleukin 2 receptor subunit beta (IL2RB) and could act as receptor of interleukin (IL)-3, IL-5, and granulocyte-macrophage colony-stimulating factor (29). PTPRC is a member of the protein tyrosine phosphatase family and regulates a number of biological activities, including proliferation, differentiation, and mitosis (14). Some previously published studies have found that PTPRC, as a kind of PTP family, could regulate the T- and B-cell antigen receptor signaling (15,16). EMP3 regulates cell growth and cell-cell interaction, and might function as a tumor suppressor in some cancers (30,31). MAPK7 as an important member of the MAPK family, and could act as a mediator for some downstream signaling pathways that are involved in a number of cellular activities, including proliferation, differentiation, transcription regulation, and development (32). Diphtheria toxin receptor (DTR) plays a pivotal role in the function of dendritic cells (DCs) (33). IGFBP2 as a secreted protein is often overexpressed in tumors and could act as a biomarker for prognosis (34). ANG and PDGFA are common mediators in the regulation of the extracellular matrix, acting as pro-oncogenic factors in the development of some cancers (35). Therefore, TNFRSF7, LTF, CSF2RB, PTPRC, and DTR are closely associated with the regulation of immune function. Moreover, EMP3, IGFBP2, ANG, and PDGFA are common regulators in the cell–cell interaction, including cell proliferation, migration, and adhesion. These results were consistent with the enrichment analysis results (Figures 2,3). The protein–protein interaction analysis showed that PTPRC was co-expressed with CSF2RB and TNFRSF7, and a significant positive correlation in gastric cancer was found, suggesting that PTPRC is a key mediator in the H. pylori infection-related gastric cancer. Therefore, PTPRC was used in the subsequent study, and the clinical value of PTPRC was evaluated. PTPRC was overexpressed in gastric cancer (Figure 6B,C and Figure 8A). The overexpression of PTPRC was positively correlated with the progression of gastric cancer (Figure 8B and Figure 9B,C). The correlation analysis also revealed that PTPRC further increased in gastric tumor tissues with H. pylori infection (Figure 9A). This result also confirmed the effect of H. pylori on PTPRC expression, indicating that PTPRC is an important gene in the process from H. pylori-related gastritis to gastric cancer. To further validate the clinical value of PTPRC on the prognosis of gastric cancer patients, a survival curve analysis of gastric cancer patients was conducted, and the data showed that the high expression of PTPRC could act as a poor prognostic factor for gastric cancer patients, especially for those at advanced stage (Figures 10,11). PTPRC is an important gene that is involved in the process of H. pylori infection, and might contribute to tumorigenesis and the development of gastric cancer.
The present study had some limitations. The sample size included in the study was small. It is necessary to increase the sample size to improve the findings of the present study. Verification of significant DEGs between H. pylori-positive or -negative gastric cancer cell lines was performed using qRT-PCR assay. Further studies, including in vivo and in vitro experiments, are needed to validate the findings of our study.
Conclusions
H. pylori-induced PTPRC is overexpressed in gastric cancer, and the overexpression of PTPRC is positively associated with the development of gastric cancer. The high expression of PTPRC could serve as a poor prognostic biomarker for gastric cancer patients, especially for those at advanced stage.
Acknowledgments
Funding: The authors would like to acknowledge Supported by Medical Scientific Research Foundation of Guangdong Province, China for financial support (A2017199).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-305
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-305
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-305). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by ethics board of Cancer Hospital Affiliated to Guangzhou Medical University. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li H, Xu CX, Gong RJ, et al. How does Helicobacter pylori cause gastric cancer through connexins: An opinion review. World J Gastroenterol 2019;25:5220-32. [Crossref] [PubMed]
- Liu XM, Xu CX, Zhang LF, et al. PBX1 attributes as a determinant of connexin 32 downregulation in Helicobacter pylori-related gastric carcinogenesis. World J Gastroenterol 2017;23:5345-55. [Crossref] [PubMed]
- de Brito BB, da Silva FAF, Soares AS, et al. Pathogenesis and clinical management of Helicobacter pylori gastric infection. World J Gastroenterol 2019;25:5578-89. [Crossref] [PubMed]
- Fulgione A, Papaianni M, Cuomo P, et al. Interaction between MyD88, TIRAP and IL1RL1 against Helicobacter pylori infection. Sci Rep 2020;10:15831. [Crossref] [PubMed]
- Khaledi M, Bagheri N, Validi M, et al. Determination of CagA EPIYA motif in Helicobacter pylori strains isolated from patients with digestive disorder. Heliyon 2020;6:e04971 [Crossref] [PubMed]
- Lee YD, Kim SE, Park SJ, et al. Efficacy of Seven-day High-dose Esomeprazole-based Triple Therapy versus Seven-day Standard Dose Non-esomeprazole-based Triple Therapy as the First-line Treatment of Patients with Helicobacter pylori Infection. Korean J Gastroenterol 2020;76:142-9. [Crossref] [PubMed]
- Irrazabal T, Thakur BK, Kang M, et al. Limiting oxidative DNA damage reduces microbe-induced colitis-associated colorectal cancer. Nat Commun 2020;11:1802. [Crossref] [PubMed]
- Nagata M, Toyonaga K, Ishikawa E, et al. Helicobacter pylori metabolites exacerbate gastritis through C-type lectin receptors. J Exp Med 2021;218:e20200815 [Crossref] [PubMed]
- Owyang SY, Zhang M, El-Zaatari M, et al. Dendritic cell-derived TGF-β mediates the induction of mucosal regulatory T-cell response to Helicobacter infection essential for maintenance of immune tolerance in mice. Helicobacter 2020;25:e12763 [Crossref] [PubMed]
- Piao JY, Kim SJ, Kim DH, et al. Helicobacter pylori infection induces STAT3 phosphorylation on Ser727 and autophagy in human gastric epithelial cells and mouse stomach. Sci Rep 2020;10:15711. [Crossref] [PubMed]
- Piao JY, Lee HG, Kim SJ, et al. Helicobacter pylori Activates IL-6-STAT3 Signaling in Human Gastric Cancer Cells: Potential Roles for Reactive Oxygen Species. Helicobacter 2016;21:405-16. [Crossref] [PubMed]
- Lv X, Zhao Y, Zhang L, et al. Development of a novel gene signature in patients without Helicobacter pylori infection gastric cancer. J Cell Biochem 2020;121:1842-54. [Crossref] [PubMed]
- Herrero R, Heise K, Acevedo J, et al. Regional variations in Helicobacter pylori infection, gastric atrophy and gastric cancer risk: The ENIGMA study in Chile. PLoS One 2020;15:e0237515 [Crossref] [PubMed]
- Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu Rev Biochem 2000;69:373-98. [Crossref] [PubMed]
- Xiu MX, Liu ZT, Tang J. Screening and identification of key regulatory connections and immune cell infiltration characteristics for lung transplant rejection using mucosal biopsies. Int Immunopharmacol 2020;87:106827 [Crossref] [PubMed]
- Goyette J, Nieves DJ, Ma Y, et al. How does T cell receptor clustering impact on signal transduction? J Cell Sci 2019;132:jcs226423 [Crossref] [PubMed]
- Wen S, Velin D, Felley CP, et al. Expression of Helicobacter pylori virulence factors and associated expression profiles of inflammatory genes in the human gastric mucosa. Infect Immun 2007;75:5118-26. [Crossref] [PubMed]
- D’Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer 2009;45:461-9. [Crossref] [PubMed]
- Cao L, Cheng H, Jiang Q, et al. APEX1 is a novel diagnostic and prognostic biomarker for hepatocellular carcinoma. Aging (Albany NY) 2020;12:4573-91. [Crossref] [PubMed]
- Xu XY, Guo WJ, Pan SH, et al. TILRR (FREM1 isoform 2) is a prognostic biomarker correlated with immune infiltration in breast cancer. Aging (Albany NY) 2020;12:19335-51. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-D613. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-W560. [Crossref] [PubMed]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580-5. [Crossref] [PubMed]
- Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016;7:49322-33. [Crossref] [PubMed]
- Starzer AM, Berghoff AS. New emerging targets in cancer immunotherapy: CD27 (TNFRSF7). ESMO Open 2020;4:e000629 [Crossref] [PubMed]
- Dhakal P, Kelleher AM, Behura SK, et al. Sexually dimorphic effects of forkhead box a2 (FOXA2) and uterine glands on decidualization and fetoplacental development. Proc Natl Acad Sci U S A 2020;117:23952-9. [Crossref] [PubMed]
- Jarjour NN, Bradstreet TR, Schwarzkopf EA, et al. BHLHE40 Promotes T(H)2 Cell-Mediated Antihelminth Immunity and Reveals Cooperative CSF2RB Family Cytokines. J Immunol 2020;204:923-32. [Crossref] [PubMed]
- Cha YJ, Koo JS. Expression and Role of Epithelial Membrane Proteins in Tumorigenesis of Hormone Receptor-Positive Breast Cancer. J Breast Cancer 2020;23:385-97. [Crossref] [PubMed]
- Thornton N, Karamatic Crew V, Tilley L, et al. Disruption of the tumour-associated EMP3 enhances erythroid proliferation and causes the MAM-negative phenotype. Nat Commun 2020;11:3569. [Crossref] [PubMed]
- Yang X, Zhong D, Gao W, et al. Conditional ablation of MAPK7 expression in chondrocytes impairs endochondral bone formation in limbs and adaptation of chondrocytes to hypoxia. Cell Biosci 2020;10:103. [Crossref] [PubMed]
- Bhave S, Arciero E, Baker C, et al. Enteric neuronal cell therapy reverses architectural changes in a novel diphtheria toxin-mediated model of colonic aganglionosis. Sci Rep 2019;9:18756. [Crossref] [PubMed]
- Wang Z, Zhao X, Ma Z, et al. Modulation on gallbladder carcinoma by TGF-β1 via IGFBP-2. Cancer Biomark 2018; Epub ahead of print. [Crossref] [PubMed]
- Li J, Diao S, Yang H, et al. IGFBP5 promotes angiogenic and neurogenic differentiation potential of dental pulp stem cells. Dev Growth Differ 2019;61:457-65. [Crossref] [PubMed]
(English Language Editor: R. Scott)


