
Identification of G-protein signaling modulator 2 as a diagnostic and prognostic biomarker of pancreatic adenocarcinoma: an exploration of its regulatory mechanisms
Introduction
Worldwide, the number of newly diagnosed cases of pancreatic cancer and the associated mortality continue to escalate every year (1). While pancreatic carcinoma is relatively rare, it is one of the most common causes of cancer mortality in Western countries (2). As invasion or distant metastasis was not rare in early stage patients, only 10–20% of patients had surgical resection of the pancreas, while the 5-year survival rate of patients undergoing surgical resection is only 23.4%, and the number drops to 6% for patients of all stages (3,4). A recent study suggested that by 2018, pancreatic cancer had be the third leading cause of cancer death in USA (5). Therefore, reducing the invasion and metastasis of pancreatic cancer has great significance for the treatment and prognosis of these patients.
G-protein signaling modulator 2 (GPSM2) belongs to a family of proteins involved in G-protein signal conditioning, and has received considerable attention in recent years (6,7). Mutations in the GPSM2 gene have been associated with poor development of the auditory system, and can lead to autosomal recessive nonsyndromic deafness (8). GPSM2 was originally found to play a vital role in ensuring the correct orientation of cell spindles and symmetric cell division (9,10). One study discovered that overexpression of GPSM2 suppressed cell growth and influenced chromosomal segregation in breast cancer (7). Pishas et al. found that inhibition of GPSM2 by XI-006, a 4-nitrobenzofuroxan derivative, had a positive therapeutic effect on Ewing sarcoma (11). Our previous studies demonstrated that GPSM2 is overexpressed in pancreatic cancer, and its expression is closely related to the T stage, TNM stage, tumor grade, and prognosis of pancreatic cancer patients (12). However, the role of GSPM2 in the growth and migration of pancreatic cancer, as well as its specific regulatory mechanism, has not been clarified.
High throughput sequencing methods are growing in popularity due to its success in early diagnosis and the prognostic evaluation of cancers (13). Since the sample size of our previous study was not large (54 samples), the results from the latter study were validated using a public database. Univariate and multivariate cox regression analyses were used to explore the factors associated with survival time in pancreatic carcinoma patients. GPSM2 was identified and shown to influence the microenvironment of pancreatic carcinomas. Since there’s no research on using microarray technology to explore the regulatory mechanism of GPSM2 at cellular whole level, stable low GPSM2 expressing pancreatic cancer cells were constructed and microarray analysis was performed to identify differentially expressed genes (DEGs). The DEGs were then examined using ingenuity pathway analysis (IPA) and protein-protein interaction (PPI) network analysis for in-depth exploration of the associated regulatory mechanisms.
We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-224).
Methods
Online analysis of gene expression and prognosis
GEPIA (Gene Expression Profiling Interactive Analysis; http://gepia.cancer-pku.cn/index.html) is a novel interactive web server developed for processing the RNA sequencing expression data of 9,736 cancers and 8,587 normal tissues obtained from the TCGA and GTEx projects by applying a standard processing pipeline (14). It has shown great promise in data analysis and presentation. The differential expression of GPSM2 between cancer tissues and normal tissues has previously been shown in a boxplot or an anthropotomical heatmap constructed using the gganatogram package (15). Preliminary survival analysis of hub genes was performed and displayed on a Kaplan-Meier curve.
TCGA data of pancreatic adenocarcinomas (PAADs)
UCSC is a data visualization and analysis website for exploring various sequencing data and related clinical phenotypic annotations from TCGA, PCAWG, Pan-Cancer Atlas, ICGC, GDC, and GTEx (16). All available TCGA data on the PAAD project were obtained from the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/). In September 2017, there were RNA sequencing data on 182 PAAD samples, which included 173 single tumor samples, 4 pairs of PAAD and adjacent non-tumor pancreatic tissues, and clinical data including survival time and survival status records of 185 patients. Prior to analysis, raw sequencing data were normalized and annotated by gene mapping. After screening, a total of 177 pancreatic cancer samples were picked up, including GPSM2 expression levels and clinical data. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Evaluation of the tumor microenvironment and immune infiltration
The amount of immunological and stromal infiltration in TCGA pancreatic carcinoma was evaluated using ESTIMATE in R software (17). The population of tumor-infiltrating immune cells in heterogeneous tissues from the transcriptomic data was quantified using the analytical platform TIMER at https://cistrome.shinyapps.io/timer (18). TIMER is a comprehensive resource for systematic analysis of immune infiltration across diverse cancer types. The infiltration abundances of six types of immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) were estimated by using a bioinformatic algorithm on the basis of rigorous statistics.
Cell culture and transfection
To induce stable low expression of GPSM2 in the PANC-1 pancreatic adenocarcinoma cell line (Institute of Cell Biology, Chinese Academy of Sciences, Shanghai, China), short hairpin (sh)RNA for GPSM2 was cloned into the GV248 vector (Genechem, Shanghai) and verified by sequencing. Three different target sequences for GPSM2 (shGPSM2#1, shGPSM2#2, shGPSM2#3), as well as the scrambled sequence as a negative control (shNC), were designed and are shown in Table S1. The PANC-1 cells were cultivated with RPMI 1640 medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) and maintained in a fully humidified atmosphere at 37 °C and 5% CO2. The culture medium was refreshed daily. Log-phase PANC-1 cells were harvested and transferred into six-well plates with serum-free medium, at a density of 5×105 cells per well. When confluency reached 70%, the cells were transfected with 4 µL of shGPSM2 or shNC sequence by applying polybrene (Genechem, Shanghai) according to the manufacturer’s protocol. The cells were transferred to complete culture medium after incubation for 16 hours. The silencing efficiency of shGPSM2 was evaluated by qPCR. Mixed populations of GPSM2-silenced cells were then used for microarray analysis and construction of an orthotopic model for future use.
Transwell migration assay
For the invasion assay, cells were cultured with serum-free medium for 24 hours after transfection. Untransfected cells (Blank group), cells transfected with the scrambled sequence (negative control, shNC group) and shGPSM2#2 sequence (shGPSM2#2 group) were then harvested and resuspended at a density of 2×105/mL in 100 µL of serum-free medium and transferred to the upper transwell chamber (Corning Costar). Next, 700 µL of complete medium containing 10% FBS was added to the lower chamber, and cells were incubated for 24 hours at 37 °C. After migration for 24 hours, the microporous membrane and the migrated cells were fixed with 4% paraformaldehyde solution for 0.5 hours and stained with 2% crystal violet (Google biotechnology, Wuhan, China) for 20 minutes. Finally, a microscope (Leica) was used for counting cells and acquiring images. Each group was repeated 3 times.
Microarray and IPA
Total RNA Isolation Reagent (SuperfecTRI) was used to extract RNA from PANC-1 cells according to the manufacturer’s protocol. The quality of RNA was verified by Agilent 2100 (Agilent), using an RNA 6000 Nano Kit (Agilent). After quality inspection, the RNA was reverse transcribed to cDNA, which was then used as a template for in vitro transcription using the GeneChip 3’ IVT Express Kit. Biotin-labeled RNA was purified and fragmented before the next step of hybridizing to Affymetrix PrimeView Human Gene Expression Array (Affymetrix) for 16 hours. The microarray was washed and stained using GeneChip Fluidics Station 450 (Affymetrix). Fluorescent images were obtained using a GeneChip Scanner 3000 (Affymetrix).
Before analysis, raw data were adjusted by mean normalization and the lowest 20% range of signal intensity of the probe was filtered out as background noise, as well as those with a coefficient of variation greater than 25% (19-21). Sequencing data were then applied to the limma package for DEGs identification based on the linear model of empirical bayesian distribution. The Benjamini-Hochberg method was applied for the correction of significant differences [false discovery rate (FDR)] (22). Genes with large variations of expression (|Fold Change| >1.5, FDR <0.05) were selected as DEGs and applied to the Ingenuity Pathways Knowledge Base (IPA Ingenuity Systems; Qiagen, Redwood City, CA) for biological information mining. According to the changes in the expression values and the correlation between them, the obtained DEGs were mapped to the predicted functional modules and shown in graph form.
Reverse transcription-quantitative PCR
The steps of RNA extraction and purification are described above. Total RNA was reverse transcribed using the RT-PCR Quick Master Mix Kit (TOYOBO, Osaka, Japan) according to the manufacturer’s protocol. After extraction, the RNA was reverse transcribed to cDNA with an oligo dT primer (Sangon Biotech, Shanghai) and M-MLV-RTase (Promega). The gene expression levels were analyzed by real-time qPCR using SYBR Green assays (TaKaRa, Otsu, Japan) on a real-time PCR System (Roche). Glyceralde-3-hydephosphate dehydrogenase (GAPDH) was used as an internal control, and the equation 2−∆∆Ct was used to describe the relative expression of the gene. The specific forward primer for GPSM2 was CCAGAGAGCTTAATGACAAGG and the reverse primer was GGACCAGGGCAACCAAAACT. The forward primer for GAPDH was TGACTTCAACAGCGACACCCA and the reverse primer was CACCCTGTTGCTGTAGCCAAA.
The PPI network and hub gene analysis
The PPI networks can analyze the specific molecular mechanisms of intracellular activity. DEGs were mapped to a PPI network by using the Search Tool for the Retrieval of Interacting Genes (STRING), which is an online database designed for visualizing PPI information (23). A confidence score of 0.4 was set as the cut-off criterion. We used the CentiScaPe plugin in Cytoscape software to calculate the node degrees, and the top 30 DEGs of node degrees were selected as hub genes (24,25). Visualization of the PPI network was also performed using Cytoscape.
Statistical analysis
IBM SPSS22.0 statistics was applied for statistical calculations. Single comparisons of measurement data between two groups were performed using the Wilcoxon Rank-Sum test, while the Kruskal-Wallis test was adopted for more groups. Survival analysis using overall survival (OS) and disease-free survival (DFS) curves was conducted according to the Kaplan-Meier method and log-rank tests were used to assess the statistical significance of differences. The prognostic significance of clinicopathologic parameters and GPSM2 expression was determined using univariate and multivariate Cox-regression analysis via a backward stepwise procedure. Differences were considered statistically significant when the P values were less than 0.05 (*P<0.05). The relative risk (RR), 95% confidence intervals (CIs), and log rank P values were computed. OS and DFS data were acquired from the clinical data of pancreatic cancer samples on the TCGA database. Graphs and curves were constructed by GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA).
Results
GPSM2 is upregulated in pancreatic adenocarcinoma (PAAD) and is correlated with clinicopathological features of PAAD
By using the GEPIA online tool (accessed on December 20, 2017), the differential expression of GPSM2 was compared between primary pancreatic carcinomas and normal samples. There was a remarkably higher expression of GPSM2 in PAAD compared to the adjacent healthy tissues (Figure 1A, P<0.001). Our previous studies confirmed this at the protein level (12). In addition, GPSM2 was overexpressed in colon adenocarcinoma, rectum adenocarcinoma, and stomach adenocarcinoma (Figure 1B, Fold Change >1), while the expression was low in thyroid carcinoma (Fold Change <1).
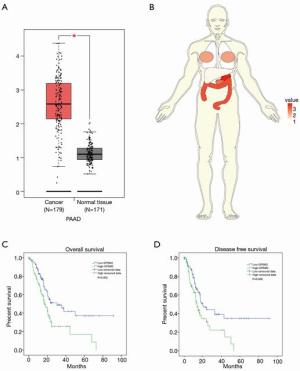
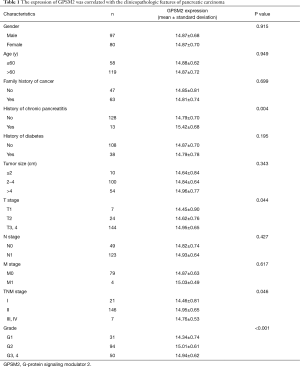
Furthermore, to determine whether GPSM2 mRNA expression levels were associated with pancreatic carcinoma progression, the relationship between GPSM2 and the clinicopathological characteristics of pancreatic carcinoma were explored by applying data from the TCGA database. The GPSM2 expression profiles of 185 samples, as well as the clinical information, were obtained from the UCSC website (https://xena.ucsc.edu/). A total of 177 PAAD samples were analyzed through rigorous selection, and the mean age of the sample cohort was 65 years (35–88 years). GPSM2 expression was markedly associated with a history of chronic pancreatitis (P=0.004), T stage (P=0.033), TNM stage (P=0.045), and tumor grade (P<0.001) (Table 1), but poorly associated with age, gender, family history of cancer, history of diabetes, tumor size, N stage, and M stage (P>0.05). These results were basically consistent with our earlier findings (12).
Full table
Association of GPSM2 expression with clinical prognosis and the tumor microenvironment
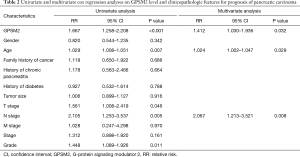
Analysis of the 177 pancreatic cancer samples showed a median OS time of 18.9 months (range, 0 to 91.4 months). A total of 92 patients died from the disease and 85 patients survived. The median DFS time was 16.4 months (range, 0 to 91.4 months). A total of 80 patients suffered a recurrence or progression of disease. Kaplan-Meier survival analysis and log-rank tests showed that patients with high GPSM2 expression suffered poorer OS and DFS compared to patients with low GPSM2 expression (Figure 1C, P=0.002; Figure 1D, P=0.008). The univariate analysis of OS in correlation with the clinicopathologic parameters and GPSM2 expression is shown in Table 2. In addition to GPSM2 protein expression (P<0.001), OS was also associated with age, T stage, N stage, and tumor grade, (P<0.05), but not with other clinicopathologic factors (P≥0.05). Multivariate analysis revealed that GPSM2 expression was an independent prognostic factor for OS in PAAD patients, in addition to age and N stage (Table 2).
Full table
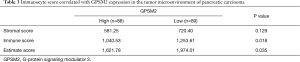
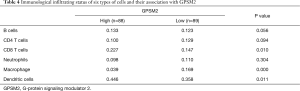
The sequencing data was further analyzed using the ESTIMATE package. Samples with higher GPSM2 expression were characterized by less dispersed infiltration of populations of immune cells (P=0.018) and stroma cells (P>0.05) (Table 3). Consistent with the lower stromal or non-tumor content, samples with higher GPSM2 expression had a higher tumor content, which can be inferred by the estimate score (Table 3, P=0.034). Immunological infiltration estimated with TIMER confirmed that tumors with lower expression of GPSM2 were characterized by a higher infiltration of macrophage cells (P<0.001), while infiltration of CD8+ T cells (P=0.01) and dendritic cells were lower (Table 4, P=0.011).
Full table
Full table
Silencing of GPSM2 expression decreased the migration ability of pancreatic cancer cells
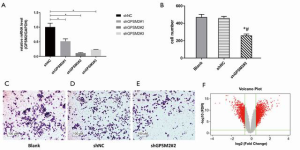
To further examine the association between GPSM2 and overall patient survival, GPSM2 was silenced in PANC-1 cells using lentiviral-mediated RNA interference (RNAi). As presented in Figure 2A, the mRNA levels of GPSM2 were decreased by 88.5% in PANC-1 cells after transfection with the shGPSM2#2 sequence (P=0.0063). To examine the biological functions of GPSM2 in PAAD cell migration, transwell assays were performed after transfecting cells with the lentiviral vector. Transwell assays demonstrated that silencing GPSM2 markedly decreased the numbers of migrating cells compared to cells transfected with the scrambled sequence (shNC) and untransfected cells (Blank group) (Figure 2B,C,D,E, P<0.001). These results suggested that GPSM2 facilitated tumor migration in pancreatic cancer cells.
Microarray analysis and identification of DEGs
Microarray analysis for the mixed population of GPSM2-silenced PANC-1 cells was performed. The gene expression profiles between cells transfected with the scrambled sequence (shNC) and shGPSM2#2 sequence were compared (Figure S1). By comparing the RNAseq read counts of the various genes and subsequently applying the cut-off criteria, 1,631 genes were identified as DEGs, including 1,039 upregulated and 592 downregulated genes. Genes with large variations of expression [Fold Change >1.5, false discovery rate (FDR) <0.05] were included in the volcano plot, while low expression genes were excluded (Figure 2F). A heatmap was painted and the DEGs in shGPSM2#2 and shNC groups were organized in obviously separate clusters (Figure S2). Comparing the mRNA expression profiles revealed that mesenchymal promoter CD44 was downregulated at the transcriptional level in the GPSM2-silenced cells, while the epithelial marker CDH1 (E-cadherin) was notably upregulated (Figure S2). These results suggested that GPSM2 promoted epithelial-to-mesenchymal transition (EMT) in PAAD cells.
IPA of the genes
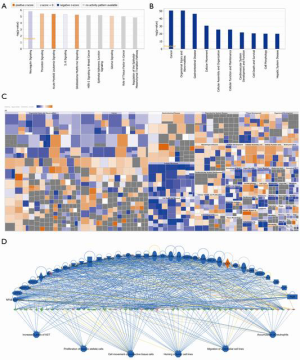
To gain further insight into the function of the identified DEGs for PAAD, enrichment analysis was performed using IPA. Several DEGs were significantly enriched in various IPA domains. In the canonical pathway, the genes were mainly enriched in neuregulin signaling, thrombin signaling, acute myeloid leukemia signaling, IL-8 signaling, and glioblastoma multiforme signaling (Figure 3A). Analysis of diseases and functions showed a focus on cancer, organismal injury and abnormalities, cellular movement, and gastrointestinal disease (Figure 3B). A heat map was constructed to examine the relationship between the expression levels of the DEGs and the activation or inhibition of the disease and function. Significantly activated diseases or functions included viral infections (Z-score =3.521), liver lesions (2.514), and others as shown in Figure 3C. Marked inhibition of disease or function involved congenital anomaly of the cardiovascular system (−3.215), congenital heart disease (−3.194), and others as shown in Figure 3C.
Analysis of upstream regulators
Upstream regulatory factors were analyzed based on the IPA dataset. Upstream regulators that were significantly activated included microRNA (miR)-124-3p, ESR1, PD98059, and MYC (Table 5). Upstream regulators that were significantly suppressed included TNF, PDGF BB, and ERK (Table 6). The top ten upstream regulators, as well as their target genes, are shown in Figure S3.

Full table

Full table
The functional implications of these upstream regulators in the development of downstream diseases and functions were investigated and several regulator effects networks were constructed (Table S2). The network with the highest consistency score (Figure 3D) suggested that upstream genes such as ALB, ATF4, C3, CD40LG, CSF1, CYR61, DICER1, EDN1, EGR1, ERK1/2, EZH2, F2, F2R, F3, FGF7, IL17A, IL36B, and integrin beta 1 (ITGB1) may inhibit accumulation of neutrophils, cell movement of connective tissue cells, homing of tumor cell lines, increased levels of AST, migration of endothelial cell lines, and proliferation of hepatic stellate cells through the downstream genes AHR, AKAP12, AKT1, ANGPTL4, ATF3, ATP2A2, AXL, CAPN2, CCL2, CD44, CD69, CXCL8, EDIL3, and EGFR.
The PPI network and the validation of hub genes
A PPI network consisting of 1,501 nodes and 8,903 edges was established by retrieving 1,631 DEGs in the STRING database (version 10.5) and visualizing through the use of Cytoscape software (Figure 4). Based on the STRING database, the DEGs with the highest PPI scores identified by the number of degrees are shown in Table S3. Among these genes, AKT1 scored maximum node degrees. The top 30 genes, including AKT1, EGFR, VEGFA, CDH1, HSP90AA1, CXCL8, KRAS, CD44, MMP2, MDM2, and ITGB1, were selected as hub genes for further analysis. The prognostic information of the 30 hub genes was freely available at http://gepia.cancer-pku.cn/index.html. The findings demonstrated that CD44 expression (Figure 5A, P=0.0051) was closely related to OS in PAAD, as was EGFR (Figure 5B, P=0.031), ITGB1 (Figure 5C, P=0.031), KRAS (Figure 5D, P=0.014), NRAS (Figure 5E, P=0.034), RAB5A (Figure 5F, P=0.029), STAT5B (Figure 5G, P=0.0016), and TFRC (Figure 5H, P=0.036).

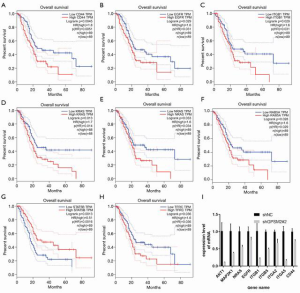
The expressions of these genes, as well as several members of the integrin family, were assessed (Figure 5I). In addition to decreased expression of CD44 (P=0.008), levels of AKT1 (P=0.023), EGFR (P=0.048), MAP2K1(P=0.033), NRAS (P=0.005), ITGB1 (P=0.021), ITGB5 (P=1.4E-04), ITGA2 (P=9.5E-03), ITGA5 (P=5.9E-04) were all significantly lower in GPSM2-silenced PNAC-1 cells compared to control cells.
Discussion
PAAD is one of the most common fatal tumors (1). This study identified GPSM2 as a new oncogene in PAAD, and revealed the correlation between GPSM2 and T stage, TNM stage, and tumor grade, which are crucial prognostic factors in PAAD. Similarly, high GPSM2 expression was also observed in several other gastrointestinal tumors, such as colon adenocarcinoma, rectum adenocarcinoma, and stomach adenocarcinoma (Figure S4). The expression of GPSM2 was low in thyroid carcinoma, which may partially explain the good prognosis in patients with thyroid carcinoma.
These findings improve our understanding of GPSM2 biology and may help clinicians predict tumor progression and prognosis, especially in patients with a high level of GPSM2 expression in the primary tumor. Furthermore, patients with tumors that exhibited a high level of GPSM2 were associated with a poor prognosis, suggesting that GPSM2 may act as a potential diagnostic and prognostic biomarker in PAAD.
With the increasing clinical success of cancer immunotherapies, there is a growing need to comprehensive understanding specific tumor immune interactions (26,27). This study demonstrated that GPSM2 could promote the infiltration of immune cells in the tumor microenvironment and reduce the content of tumor cells. Dendritic cells have been reported to internalize antigens from the tumor and process them to present them to CD8+ T cells (28). Also, macrophages play large roles in the promotion of pancreatic tumor growth and development of an immunosuppressive microenvironment (29). Our study found that GPSM2 expression is associated with the infiltration of macrophage cells, CD8+ T cells and dendritic cells, suggesting that the function of GPSM2 in pancreatic cancer may be related to these components.
It has been reported that G-protein coupled receptors (GPCRs) play a critical role in the enhancement of breast cancer cell growth (7). However, it remained to be determined whether GPSM2 is also involved in the carcinogenesis of the pancreas. The transwell assays in this investigation revealed that GPSM2 facilitated cell migration in PANC-1 cells.
In this analysis, 1,631 DEGs were identified, consisting of 1,039 upregulated genes and 592 downregulated genes. The canonical pathways analysis showed that these DEGs were involved in epithelial adherens junction signaling and regulation of the EMT pathway. There is increasing evidence that EMT represents the key cellular reprogramming required for metastasis and chemoresistance in PAAD (30). Several growth factors have been documented to trigger EMT (31,32). A recent study reported that epithelial cadherin (CDH1) is involved in mechanisms regulating cell-cell adhesions, mobility, and proliferation of epithelial cells (33).
This investigation demonstrated that the expressions of the EMT-related proteins CD44 and SNAII2 were decreased, while E-cadherin was increased in the shRNA cells (Figure 5I), suggesting that GPSM2 is a novel EMT inducer in PAAD.
Some reports have suggested that the PI3K/AKT pathway that transmits signals downstream of GPCRs is dysregulated in breast cancer (34). Another study suggested that GPSM2 may be a critical molecule for carcinoma progression and may act as a potential biomarker and therapeutic target for the prevention and treatment of PAAD (6). The present study found that GPSM2 could regulate AKT1, and this may be the mechanism by which GPSM2 promotes migration of pancreatic carcinoma cells. Integrins are cell-adhesion molecules, and reports have shown that its upregulation is associated with tumor progression, migration, invasion, and metastasis (7,35).
Studies have demonstrated that ITGB1 can promote the growth and migration period of tumors in many types of human cancers, including pancreatic cancer (36-39), and thus ITGB1 may be a therapeutic target for the treatment of these cancers. In fact, a monoclonal antibody against ITGB1 has been shown to block hepatocellular cancer cell invasion (40) and knockdown of ITGB1 by lentivirus-based RNAi inhibited PC cell growth and migration in vitro and in vivo (38). Indeed, the results of this current study suggested that ITGB1 may be a downstream gene of GPSM2 involved in oncogenesis.
In conclusion, this investigation identified GPSM2 as a potential diagnostic and prognostic biomarker and its specific regulation mechanisms were preliminarily explored. Future studies should include gain-of-function experiments and in vivo experiments using xenografts in nude mice to further examine the effects of GPSM2 in PAAD. In addition, the detailed mechanisms of GPSM2 and its downstream pathways warrant further investigation. Finally, this study suggested that GPSM2 may be a critical molecule for carcinoma progression and may be a potential biomarker and therapeutic target for the prevention and treatment of PAAD.
Acknowledgments
We thank LetPub (
Funding: This work was supported by the Key Research and Development Project of Zhenjiang (Social Development Project; SH2019061).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-224
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-224). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol 2009;6:699-708. [Crossref] [PubMed]
- Sener SF, Fremgen A, Menck HR, et al. Pancreatic cancer: a report of treatment and survival trends for 100,313 patients diagnosed from 1985-1995, using the National Cancer Database. J Am Coll Surg 1999;189:1-7. [Crossref] [PubMed]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. [Crossref] [PubMed]
- He XQ, Zhang YF, Yu JJ, et al. High expression of G-protein signaling modulator 2 in hepatocellular carcinoma facilitates tumor growth and metastasis by activating the PI3K/AKT signaling pathway. Tumour Biol 2017;39:1010428317695971 [Crossref] [PubMed]
- Fukukawa C, Ueda K, Nishidate T, et al. Critical roles of LGN/GPSM2 phosphorylation by PBK/TOPK in cell division of breast cancer cells. Genes Chromosomes Cancer 2010;49:861-72. [Crossref] [PubMed]
- Doherty D, Chudley AE, Coghlan G, et al. GPSM2 mutations cause the brain malformations and hearing loss in Chudley-McCullough syndrome. Am J Hum Genet 2012;90:1088-93. [Crossref] [PubMed]
- Williams SE, Beronja S, Pasolli HA, et al. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 2011;470:353-8. [Crossref] [PubMed]
- Du Q, Macara IG. Mammalian pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 2004;119:503-16. [Crossref] [PubMed]
- Pishas KI, Adwal A, Neuhaus SJ, et al. XI-006 induces potent p53-independent apoptosis in Ewing sarcoma. Sci Rep 2015;5:11465. [Crossref] [PubMed]
- Peng Y, Cui L, Shi J, et al. Expression of G-protein signaling modulator 2 in pancreatic cancer tissues. Jiangsu Daxue Xuebao Yixueban 2016;26:231-4.
- Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol 2008;5:588-99. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Maag JLV. gganatogram: an R package for modular visualisation of anatograms and tissues based on ggplot2. F1000Res 2018;7:1576. [Crossref] [PubMed]
- Goldman M, Craft B, Hastie M, et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. 2019 [cited 2019 July 24]. Available online:
10.1101/326470 10.1101/326470 - Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17:174. [Crossref] [PubMed]
- Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185-93. [Crossref] [PubMed]
- Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003;31:e15 [Crossref] [PubMed]
- Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249-64. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47 [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Scardoni G, Tosadori G, Faizan M, et al. Biological network analysis with CentiScaPe: centralities and experimental dataset integration. F1000Res 2014;3:139. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Sharma P, Wagner K, Wolchok JD, et al. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat Rev Cancer 2011;11:805-12. [Crossref] [PubMed]
- Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1-10. [Crossref] [PubMed]
- Cui R, Yue W, Lattime EC, et al. Targeting tumor-associated macrophages to combat pancreatic cancer. Oncotarget 2016;7:50735-54. [Crossref] [PubMed]
- Maier HJ, Wirth T, Beug H. Epithelial-mesenchymal transition in pancreatic carcinoma. Cancers (Basel) 2010;2:2058-83. [Crossref] [PubMed]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178-96. [Crossref] [PubMed]
- Biamonti G, Bonomi S, Gallo S, et al. Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT). Cell Mol Life Sci 2012;69:2515-26. [Crossref] [PubMed]
- Meigs TE, Fedor-Chaiken M, Kaplan DD, et al. Gα12 and Gα13 negatively regulate the adhesive functions of cadherin. J Biol Chem 2002;277:24594-600. [Crossref] [PubMed]
- Ye Y, Tang X, Sun Z, et al. Upregulated WDR26 serves as a scaffold to coordinate PI3K/AKT pathway-driven breast cancer cell growth, migration, and invasion. Oncotarget 2016;7:17854-69. [Crossref] [PubMed]
- Gotzmann J, Mikula M, Eger A, et al. Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutat Res 2004;566:9-20. [Crossref] [PubMed]
- Caccavari F, Valdembri D, Sandri C, et al. Integrin signaling and lung cancer. Cell Adh Migr 2010;4:124-9. [Crossref] [PubMed]
- Felding-Habermann B. Integrin adhesion receptors in tumor metastasis. Clin Exp Metastasis 2003;20:203-13. [Crossref] [PubMed]
- Grzesiak JJ, Cao HST, Burton DW, et al. Knockdown of the β1 integrin subunit reduces primary tumor growth and inhibits pancreatic cancer metastasis. Int J Cancer 2011;129:2905-15. [Crossref] [PubMed]
- Song J, Zhang J, Wang J, et al. β1 integrin mediates colorectal cancer cell proliferation and migration through regulation of the Hedgehog pathway. Tumour Biol 2015;36:2013-21. [Crossref] [PubMed]
- Masumoto A, Arao S, Otsuki M. Role of β1 integrins in adhesion and invasion of hepatocellular carcinoma cells. Hepatology 1999;29:68-74. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


