
Profiles of immune infiltration in the tumor microenvironment of hepatocellular carcinoma
Introduction
Liver cancer is the sixth most common malignant tumor and the fourth leading cause of death from tumors in the world. In 2018, there were 841,080 new cases of liver cancer, with 781,631 patients dying from this disease (1). Hepatocellular carcinoma (HCC) is the most common type of liver cancer. The main treatment strategies for HCC are surgery, radiotherapy, chemotherapy, and palliative therapies, such as radiofrequency ablation (2). However, the efficacy of these treatments in most patients with recurrence or distant metastasis is limited (3). Therefore, it is necessary to explore a new feasible treatment strategy for HCC.
Tumor microenvironment (TME) plays a crucial role in the growth, metastasis, and prognosis of tumors (4,5). The TME-infiltrating immune cells (TIICs) are immunocytes that transfer from peripheral blood to tumor tissue and are closely related to the antitumor response (6,7). The number of immune cells varies from tissue to tissue, so the different tissues in hepatocellular carcinoma have different immune response. TIICs are major players in promoting anti-cancer immune responses, and are recognized as a crucial prognostic factor of the patients with malignant tumors, especially since their levels can predict therapeutic effectiveness and survival, so it have garnered increased attention in recent research (8-10).
The existing experimental methods of immunohistochemical and flow cytometry for detecting TIICs cannot investigate the diversity of immune cells infiltrating the same tissue sample at the same time and cannot comprehensively evaluate the role of immune cells in TME. Therefore, cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT), an algorithm that uses gene expression data from bulk tumor to accurately calculate the abundance of various immunocyte types infiltrating the TME, has been widely used to evaluate the abundance of 22 immune cell types (11). Through this technology, we can obtain a huge number of Gene expression data at once and allowing us to identify prognostic and therapeutic markers based on immune cells. Research published in the past few years has mainly focused on the abundance of TIICs in HCC, but there have been no further comprehensive analyses (12). However, systematic studies on immune subtypes of HCC and the degree of corresponding immune cell infiltration, survival analysis, and the analysis of immunoscore have not been explored. Thus, the comprehensive profile of immune cell infiltration of the TME in HCC has still not been fully elucidated. Therefore our study aimed to comprehensively characterize tumor-infiltrating immune cells (TIICs) and immune subtypes in HCC to identify associations with the prognosis of HCC patients.
Therefore, in the present study, we used the transcriptional sequence samples of 371 patients with HCC from The Cancer Genome Atlas (TCGA) database, employed the CIBERSORT algorithm to estimate the proportion of 22 immune cell types in HCC, and used unsupervised cluster analysis to define the immunological subtypes of HCC. We further investigated the relationship of immune subtypes with the clinical outcome, immunomodulator (IM) gene expression, programmed cell death protein 1/programmed death-ligand 1 (PD-1/PD-L1) expression, and the interaction network of immunocytes. In addition, the immune scores of HCC patients and their relationship with survival rates were evaluated. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-291).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data sources and processing
The RNA-sequencing data and corresponding clinical follow-up information of patients with HCC were downloaded from TCGA database (https://cancergenome.nih.gov/). Patients without clinical information were removed. Finally, 371 patients with HCC were included in this study.
Abundance of TME-infiltrating immune cells in HCC
To elucidate the immune infiltration landscape of HCC, the CIBERSORT algorithm was used to estimate the relative abundance of 22 human immune cell types. The CIBERSORT algorithm can evaluate the infiltration proportions of immunocyte types in tumor tissues by using the LM22 gene signature based on deconvolution (11). We loaded 371 eligible RNA-sequencing data of TCGA samples into the CIBERSORT website (https://ciber fortstanford.edu/), and the threshold value was set to a P value <0.05. Then, we obtained the Iscores (Immune Score) of 22 immune cells, and further studies were performed.
Analysis of HCC immune subtypes
According to the 5 representative immune gene sets, including macrophages (13), interferon gamma (IFN-γ) (14), transforming growth factor (TGF-β) (15), wound-healing (16), and leukocyte infiltration (17), single-sample geneset enrichment analysis (ssGSEA) was performed to analyze the enrichment of immune cells and calculate the ssGSEA score (18,19). Then, the expectation-maximization (EM) algorithm from the “mclust” R package (The R Foundation for Statistical Computing) was used to perform unsupervised cluster analysis on the results of ssGSEA. The K value corresponding to the maximum Bayesian Information Criterion (BIC) was selected, and finally, the HCC was categorized into three immune subtypes.
Consistency index
The consistency index (C-index) was calculated to compare the accuracy of the predicted value of the Cox regression model. With survival analysis, for example, if the predicted survival rate of one patient with a longer survival time is higher than that of the other, this indicates that the predicted value is consistent with the real value, which is called consistency. The normal value of the C-index is between 0.50 and 1.00. In general, the value of the C-index is in the range between 0.50 and 0.70, indicating that the accuracy of the predicted value of the Cox regression model is low. The value of the C-index that is in the range of 0.70 to 0.90 indicates medium accuracy, while a value higher than 0.90 represents high accuracy.
Tumor immune score
According to the abundance of immune cells in HCC estimated by the CIBERSORT algorithm, the immune scores were calculated by using the prognosis-related regression models constructed by Zeng et al. in gastric cancer samples and Li et al. in liver cancer samples (20,21). The thresholds of prognosis-related immune score were –0.37 and 0.10, respectively. All patients with HCC were divided into a high immunoscore group and a low immunoscore group. Then, the Kaplan-Meier analysis was performed to evaluate the survival rates of patients with different immune scores.
Principal component analysis (PCA)
PCA is a mathematical dimensionality reduction method, which uses an orthogonal transformation to transform a series of linearly related variables to a new set of linearly unrelated variables (also called the principal component). These new variables are then used to show the characteristics of the data in a smaller dimension.
The principal component is a linear combination of the original variables, and the number of principal components is not more than the number of original variables. After the combination, we obtained a set of new observation data. The meaning of these new data was different from that of the original data. These new data not only contain most of the characteristics of the previous data but also have a lower dimension, which is convenient for further analysis.
Statistical analysis
The Kaplan-Meier method was used to draw the survival curves, while the log-rank test was used to determine whether the differences were statistically significant. Continuous variables are shown as mean ± SD and were examined by t test or Kruskal-Wallis test. All statistical analyses were conducted using R v.3.6.2 software (https://www.r-project.org/). The P values were 2-sided, and a P value <0.05 was considered statistically significant.
Results
The abundance of 22 TME-infiltrating immune cells types in HCC
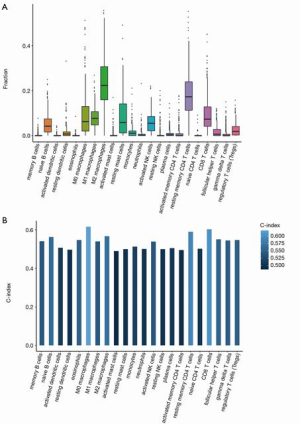
TIICs are associated with the clinical outcome of patients and are likely to be used as immunotherapy targets to improve the survival rate of patients with tumors. We used the CIBERSORT algorithm to estimate the abundance of TIICs in HCC. The results of the CIBERSORT analysis revealed that macrophages (M0, M1, and M2), resting mast cells, resting memory CD4 T cells, and CD8 T cells represented a greater proportion of infiltration compared with other immunocytes types (Figure 1A). Furthermore, C-index was used to analyze the effects of 22 immune cell types on the prognosis of patients with HCC. The results showed that C-index values of 22 TIICs were all less than 0.7, indicating that all TIICs had little predictive effect on the prognosis of patients with HCC (Figure 1B).
The immune subtypes of TME in HCC
To further explore the immunological subtypes of immune cell infiltration in HCC, cluster analysis was performed according to five immune gene sets (macrophages, IFN-γ, TGF-β, wound healing, and leukocyte infiltration). The cluster analysis revealed three distinct immune subtypes of HCC. Immune subtype 1 (C1 subtype) was characterized by increases in the expression of macrophages, IFN-γ, TGF-β, wound healing, and leukocyte infiltration. Immune subtype 2 (C2 subtype) exhibited a high expression of TGF-β and wound healing. Immune subtype 3 (C3 subtype) showed significant increases in IFN-γ expression (Figure 2A).
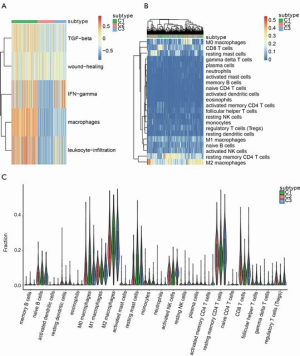
To investigate whether there are differences in immune cell types and immune activation among different immune subtypes, we conducted unsupervised cluster analysis for all samples according to immune cell types and the relative abundance of these immunocytes. It was found that C1 subtype was characterized by increases in the infiltration of macrophages (M0 and M2), resting mast cells, resting memory CD4 T cell, and CD8 T cells (Figure 2B). We further calculated the abundance of TIICs in the three different immune subtypes using the CIBERSORT algorithm (Figure 2C), and the results were consistent with those depicted in Figure 2B.
Survival analysis of immune subtypes of HCC
To further explore the relationship between immune subtypes of HCC and the prognosis of patients, we used the Kaplan-Meier method to analyze the survival rates. This revealed that the survival time of the C1 subtype and C3 subtype was longer than that of the C2 subtype, although there was no significant difference in survival rate among the three distinct immune subtypes (P>0.05; Figure 3A).
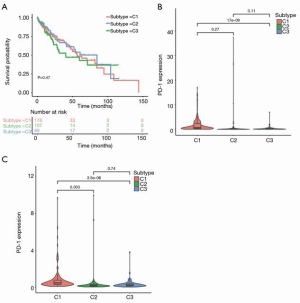
Immune checkpoint inhibitors (ICIs), such PD-1 and PD-L1 are important immunosuppressive molecules. Accumulating evidence indicates that the level of PD-1/PD-L1 expression is closely related to the antitumor immune response, and immunotherapy based on the PD-1/PD-L1 has shown remarkable success in some patients. Therefore, we analyzed the expression of PD-1/PD-L1 in these immune subtypes and found that increased expression of PD-1/PD-L1 was associated significantly with different immune subtypes (P<0.05; Figure 3B,C). This indicated that the C1 subtype may be more responsive to immunotherapy based on ICIs, which may also explain the relatively higher survival rate of the patients with the C1 subtype.
The expression of immunomodulator genes in the three distinct immune subtypes
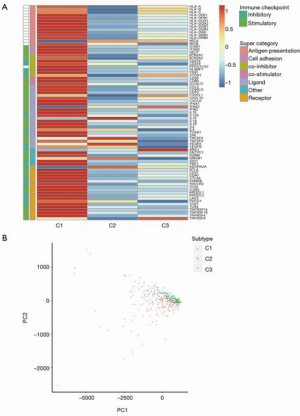
Immunomodulators (IMs) are essential for tumor immunotherapy (22), and many clinical trials have been carried out to evaluate the efficacy of IM agonists and antagonists in tumor patients. Therefore, it was particularly important to analyze the differences in the expression of IM genes in three distinct immune subtypes, so as to select suitable IM agonists and antagonists for further antitumor immunotherapy. By estimating the IM gene expression of the patients with HCC, we found that there were significant differences in the expression level of IM genes in different immune subtypes (Figure 4A). The expression of IM genes in the C1 subtype was significantly higher than that in the C2 subtype and C3 subtype. Further PCA of IM gene expression showed that PCA, to some extent, could also distinguish between different immune subtypes of HCC (Figure 4B).
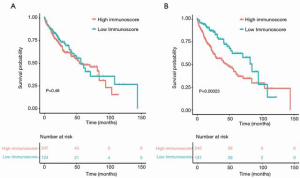
The survival analysis of patients with different tumor immune scores
Previous studies have shown TME score to be a prognostic biomarker that could predict immunotherapeutic efficacy (23). Therefore, we calculated the prognosis-related immune scores of patients with HCC according to the relative abundance of 22 kinds of immune cells. Subsequently, we divided 371 HCC patients into a high immunoscore group and a low immunoscore group to investigate the relationship between survival probability and immune score. The results showed that the survival rate of patients with low immune score was higher than that of patients with a high immune score (as calculated by the regression model of Zeng et al.), but this difference was not statistically significant (P>0.05; Figure 5A). The survival time of patients with low immunoscore was longer compared that of the high immunoscore group (according to the regression model constructed by Li et al.) and the difference was statistically significant (P<0.01, Figure 5B).
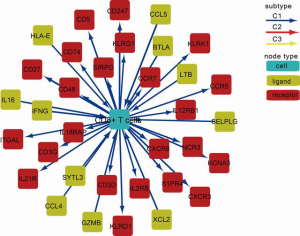
Analysis of the interaction network of immune cells in HCC
The communication relationship between immune and nonimmune stromal cells in different TMEs is very complex. By analyzing the interaction of immune cells in tumor tissues, we may be able to find a significantly enriched cellular interaction network in different immune subtypes that may play a guiding role in tumor immunotherapy. Figure 6 shows the interaction network of CD8+ T cells, indicating that CD8+ T cell communication was mainly concentrated in the C1 subtype, reflecting the immune activation of the C1 subtype, which was consistent with the results of the previous analysis of the relative abundance of TIICs and immune subtypes in HCC.
Discussion
Analysis of the profiles of tumor immune infiltration can be used to predict the clinical outcome of tumor patients and provide a more effective strategy for tumor immunotherapy. Although Rohr-Udilova et al. analyzed the fraction of TIICs in HCC in detail and completed a prognostic correlation analysis (12), there have been no further comprehensive analyses of TIICs in HCC. Therefore, on the basis of estimating the relative abundance of TIICs in HCC, we further analyzed the expression of genes related to the prognosis and immunotherapy responsiveness of patients. The interaction network of immunocytes was also investigated, and the immune scores of HCC patients were calculated to provide an even more comprehensive analysis.
The CIBERSORT analysis of TIICs showed that there were more tumor-infiltrating macrophages in HCC and that the macrophages (M0 and M2) were mainly enriched in the C1 subtype. However, several studies have shown that tumor-associated macrophages are able to promote the growth of liver cancer by promoting angiogenesis, and the infiltration of macrophages in tumor tissue is closely related to the poor prognosis of patients (24,25). Our results are not consistent with previous studies, and this should be clarified by further experiments and clinical trials. Moreover, resting mast cells were also abundant in HCC and significantly enriched in the C1 subtype. Since mast cells are the key regulatory cells of immune effector cells, activating mast cells is likely to provide a treatment target for immunotherapy. Recently, studies by Fleischmann et al. have shown that a high density of mast cells in prostate cancer is associated with good prognosis (26). Another study has demonstrated that activated mast cells have a protective effect on the development of tumors (27). Therefore, inactivating mast cells in HCC may promote tumor immune escape, thus facilitating tumor growth. The results of our study verified the previous research results. In addition, it has been reported that there is high infiltration of CD8+ T cells in the tissue of HCC and that this CD8+ T cell infiltration is an independent prognostic factor for patient outcome (28-30). The results of interaction network analysis of the present study also showed that CD8+ T cell communication was mainly concentrated in the C1 subtype, reflecting the immune activation of the C1 subtype, which is consistent with the results of previous studies.
IMs are substances that can regulate the immune system and activate or inhibit the immune response, and include immune stimulators and immunosuppressants, which can be used in antitumor immunotherapy (31). We found that the expression of IM genes in the C1 subtype was higher than that in the other two subtypes, indicating that the C1 subtype might have a good response to immunotherapy based on IMs.
The application of immunotherapy based on ICIs has completely changed the clinical landscape of tumor treatment strategies and greatly improved the survival of patients with advanced cancer. Immune checkpoint blockade plays an important role in regulating TME and antitumor immune response. ICIs are mainly expressed on the surface of activated T cells and can initiate the programmed death of T cells by binding to corresponding specific ligands. At present, ICIs have been approved for the clinical treatment of various tumors, including pancreatic cancer, lung cancer, breast cancer, and others (32-34). Our results showed that the expression of PD-1/PD-L1 in the C1 subtype was significantly higher than that in the other two immune subtypes, indicating that the C1 subtype would be more responsive to ICIs, thus providing evidence for anti-HCC immunotherapy based on ICIs.
By calculating the immune score, the infiltration of immune cells in HCC can be judged accurately. The results of this study suggested that the prognosis of the patients with a low immunoscore was better than that of patients with a high immunoscore. However, previous studies have shown that the high immune score of liver cancer is related to a better outcome (35-37), which is not in line with findings of the present study. This discrepancy may be attributable to the fact that the scoring system used in our study to calculate the immune score was different from those used in prior studies. Therefore, a more reasonable algorithm for calculating the immune score needs to be developed. Further research is also needed to elucidate this inconsistency. Nonetheless, immune score has potential as a prognostic marker for HCC and could help clinicians better understand the immune status of patients.
Taken together, the evaluation of TIICs in HCC patients and the definition of immune subtypes of HCC provided useful predictors for clinical outcome. The C1 subtype of HCC may show better immunoactivity in antitumor immunotherapy. However, there is a limitation in our study that should be specifically mentioned. The results of our research were obtained from mining data that had not been fully verified by experiments and clinical trials. Therefore, the role of TIICs in HCC should be further validated in future studies.
Acknowledgments
Funding: This study was funded by Natural Science Basic Research Program of Shaanxi (Program No. 2021JM-256).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-291
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-291). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Llovet JM, Villanueva A, Lachenmayer A, et al. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 2015;12:408-24. [Crossref] [PubMed]
- Tian G, Yang S, Yuan J, et al. Comparative efficacy of treatment strategies for hepatocellular carcinoma: systematic review and network meta-analysis. BMJ Open 2018;8:e021269 [Crossref] [PubMed]
- Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306. [Crossref] [PubMed]
- Yang JD, Nakamura I, Roberts LR. The tumor microenvironment in hepatocellular carcinoma: current status and therapeutic targets. Semin Cancer Biol 2011;21:35-43. [Crossref] [PubMed]
- Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018;68:1025-41. [Crossref] [PubMed]
- Chen QF, Li W, Wu PH, et al. Significance of tumor-infiltrating immunocytes for predicting prognosis of hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol 2019;25:5266-82. [Crossref] [PubMed]
- Jang JE, Hajdu CH, Liot C, et al. Crosstalk between Regulatory T Cells and Tumor-Associated Dendritic Cells Negates Anti-tumor Immunity in Pancreatic Cancer. Cell Rep 2017;20:558-71. [Crossref] [PubMed]
- Miksch RC, Schoenberg MB, Weniger M, et al. Prognostic impact of tumor-infiltrating lymphocytes and neutrophils on survival of patients with upfront resection of pancreatic cancer. Cancers (Basel) 2019;11:39. [Crossref] [PubMed]
- Calderaro J, Petitprez F, Becht E, et al. Intra-tumoral tertiary lymphoid structures are associated with a low risk of early recurrence of hepatocellular carcinoma. J Hepatol 2019;70:58-65. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Rohr-Udilova N, Klinglmuller F, Schulte-Hermann R, et al. Deviations of the immune cell landscape between healthy liver and hepatocellular carcinoma. Sci Rep 2018;8:6220. [Crossref] [PubMed]
- Beck AH, Espinosa I, Edris B, et al. The macrophage colony-stimulating factor 1 response signature in breast carcinoma. Clin Cancer Res 2009;15:778-87. [Crossref] [PubMed]
- Wolf DM, Lenburg ME, Yau C, et al. Gene co-expression modules as clinically relevant hallmarks of breast cancer diversity. PLoS One 2014;9:e88309 [Crossref] [PubMed]
- Teschendorff AE, Gomez S, Arenas A, et al. Improved prognostic classification of breast cancer defined by antagonistic activation patterns of immune response pathway modules. BMC Cancer 2010;10:604. [Crossref] [PubMed]
- Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol 2004;2:E7 [Crossref] [PubMed]
- Calabrò A, Beissbarth T, Kuner R, et al. Effects of infiltrating lymphocytes and estrogen receptor on gene expression and prognosis in breast cancer. Breast Cancer Res Treat 2009;116:69-77. [Crossref] [PubMed]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12. [Crossref] [PubMed]
- Scrucca L, Fop M, Murphy TB, et al. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J 2016;8:289-317. [Crossref] [PubMed]
- Zeng D, Zhou R, Yu Y, et al. Gene expression profiles for a prognostic immunoscore in gastric cancer. Br J Surg 2018;105:1338-48. [Crossref] [PubMed]
- Li W, Xu L, Han J, et al. Identification and validation of tumor stromal immunotype in patients with hepatocellular carcinoma. Front Oncol 2019;9:664. [Crossref] [PubMed]
- Chen YL, Chang MC, Cheng WF. Metronomic chemotherapy and immunotherapy in cancer treatment. Cancer Lett 2017;400:282-92. [Crossref] [PubMed]
- Zeng D, Li M, Zhou R, et al. Tumor Microenvironment Characterization in Gastric Cancer Identifies Prognostic and Immunotherapeutically Relevant Gene Signatures. Cancer Immunol Res 2019;7:737-50. [Crossref] [PubMed]
- Ding W, Tan Y, Qian Y, et al. Clinicopathologic and prognostic significance of tumor-associated macrophages in patients with hepatocellular carcinoma: A meta-analysis. PLoS One 2019;14:e0223971 [Crossref] [PubMed]
- Yeung OW, Lo CM, Ling CC, et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol 2015;62:607-16. [Crossref] [PubMed]
- Fleischmann A, Schlomm T, Köllermann J, et al. Immunological microenvironment in prostate cancer: high mast cell densities are associated with favorable tumor characteristics and good prognosis. Prostate 2009;69:976-81. [Crossref] [PubMed]
- Singer J, Jensen-Jarolim E. IgE-based Immunotherapy of Cancer -A Comparative Oncology Approach. J Carcinog Mutagen 2014;5:1000176 [Crossref] [PubMed]
- Huang CY, Wang Y, Luo GY, et al. Relationship Between PD-L1 Expression and CD8+ T-cell Immune Responses in Hepatocellular Carcinoma. J Immunother 2017;40:323-33. [Crossref] [PubMed]
- Flecken T, Schmidt N, Hild S, et al. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology 2014;59:1415-26. [Crossref] [PubMed]
- Gabrielson A, Wu Y, Wang H, et al. Intratumoral CD3 and CD8 T-cell densities associated with relapse-free survival in HCC. Cancer Immunol Res 2016;4:419-30. [Crossref] [PubMed]
- Lavelle EC, McLachlan JB. Editorial overview: Immunomodulation: Striking the right balance: using immunomodulators to target infectious diseases, cancer, and autoimmunity. Curr Opin Pharmacol 2018;41:vii-ix. [Crossref] [PubMed]
- Feng M, Xiong G, Cao Z, et al. PD-1/PD-L1 and immunotherapy for pancreatic cancer. Cancer Lett 2017;407:57-65. [Crossref] [PubMed]
- Schütz F, Stefanovic S, Mayer L, et al. PD-1/PD-L1 Pathway in Breast Cancer. Oncol Res Treat 2017;40:294-7. [Crossref] [PubMed]
- Sui H, Ma N, Wang Y, et al. Anti-PD-1/PD-L1 Therapy for Non-Small-Cell Lung Cancer: Toward Personalized Medicine and Combination Strategies. J Immunol Res 2018;2018:6984948 [Crossref] [PubMed]
- Wei L, Delin Z, Kefei Y, et al. A classification based on tumor budding and immune score for patients with hepatocellular carcinoma. Oncoimmunology 2019;9:1672495 [Crossref] [PubMed]
- Ge PL, Li SF, Wang WW, et al. Prognostic values of immune scores and immune microenvironment-related genes for hepatocellular carcinoma. Aging (Albany NY) 2020;12:5479-99. [Crossref] [PubMed]
- Yao Q, Bao X, Xue R, et al. Prognostic value of immunoscore to identify mortality outcomes in adults with HBV-related primary hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e6735 [Crossref] [PubMed]
(English Language Editor: J. Gray)


