
TSC1/2 mutations—a unique type of mutation suitable for liver transplantation of Hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the fourth most common malignant tumor globally (1) and ranks second among cancer deaths in China. Half of the total number of new liver cancer cases and deaths in China are attributed to HCC (2), indicating that HCC prevention and control are particularly challenging. For patients with advanced HCC, liver transplantation is currently the best treatment method. However, HCC recurrence after transplantation is a common complication (3-5), affecting the survival rate and prognosis of patients and posing challenges for medical providers. Adjuvant chemotherapy does not affect the recurrence rate of HCC (6), and traditional chemotherapy drugs have toxic side effects and cause liver damage, and many patients with HCC exhibit primary resistance. Therefore, traditional chemotherapy is not an ideal treatment for HCC that recurs after transplantation, nor does it improve the patient survival rate (7-10). Although two new drugs for molecular targeted therapy, sorafenib, and regorafenib, are effective for advanced HCC and HCC that recurs after transplantation, they only prolong the median patient survival by 2–3 months (11-13). After liver transplantation, HCC patients must take immunosuppressants for life, balance immunosuppression, and anti-tumor immunity is also an issue.
The key pathogenic factors involved in the tumor-causing process in HCC are WNT/β-catenin, Hedgehog, hepatocellular growth factor/c-MET, vascular endothelial growth factor (VEGF), mitogen-activated protein kinase (MAPK)/ERK (or Ras) -Raf-MEK-ERK), and PI3K/AKT/mTOR. The mammalian target of the sirolimus (mTOR) pathway is particularly important because its mutation is closely related to the efficacy of sirolimus in patients with HCC who undergo liver transplantation, and it is associated with a more aggressive tumor progression and shorter survival. Recent studies have shown that the phosphatidylinositol 3 kinase (PI3K)/protein kinase B (PKB/AKT)/(mTOR) signaling pathway plays a key role in the pathogenesis of HCC (14). Guertin et al. (15) used sirolimus to reveal the process of mTOR dependence, suggesting that mTOR regulates cell growth by controlling mRNA translation, ribosome biogenesis, autophagy, and metabolism. After liver transplantation for HCC, treatment with sirolimus, an mTOR pathway inhibitor, had both immunosuppressive effects and inhibited cancer cells’ growth and metastasis (15,16). Studies have shown that sirolimus is superior to other types of immunosuppressants for reducing tumor recurrence after liver transplantation for HCC (17,18). However, there is currently no unified opinion in the academic community on whether sirolimus can improve the long-term survival rate and prognosis of patients who undergo liver transplantation for HCC (19-21). Previous studies have not reported the use of gene detection in patients with liver transplantation for determining precise medication to reduce the recurrence of HCC after liver transplantation. Also, existing studies have not systematically combined clinical data to investigate a link between mTOR-related gene mutations and sirolimus efficacy. As many factors affect the prognosis of patients with HCC who undergo liver transplantation, debates exist on patients’ long-term prognosis, and it remains unclear whether all patients with HCC who undergo liver transplantation will benefit from sirolimus treatment. In the context of precision medicine, as the cost of whole-genome sequencing has fallen sharply, it has become a general trend to understand the genotype of each patient undergoing liver transplantation for HCC at the genetic level and develop personalized transplantation immunity and anti-tumor immunity regimens. Therefore, our study examined the mTOR-related genes of patients after liver transplantation for HCC as a means of determining personalized and precise medicine regimes in these patients.
We present the study in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-378).
Methods
Patients and settings
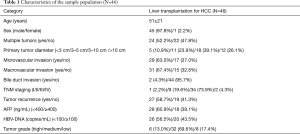
We retrospectively collected and analyzed data of 46 patients who underwent liver transplantation for HCC at the Third Affiliated Hospital of Sun Yat-Sen University, China, from December 2012 to January 2018. The patients did not receive radiotherapy, chemotherapy, and/or immunosuppressive therapy before surgery. All patients received routine treatment with basiliximab, a glucocorticoid withdrawal regimen, and tacrolimus immunosuppressive therapy. The data collected mainly included the parameters related to tumor recurrence or patient survival time after transplantation, including the demographic data of the patient (age, sex, etc.), the relevant condition of the tumor before transplantation (primary tumor size, presence, or absence of blood vessel invasion, etc.), and the postoperative information (tumor pathology, postoperative anti-rejection regimen, tumor recurrence, etc.) of each patient. The inclusion criteria included all of the following: (I) patients who underwent allogeneic orthotopic liver transplantation; (II) patients with a confirmed diagnosis of HCC; (III) patients who gave informed consent and agreed to participate in this study; and (IV) patients who completed the scheduled follow-up.
The exclusion criteria were any of the following: (I) patients with incomplete clinical data; (II) patients with poor compliance; (III) patients with diseases such as HIV and other malignant tumors. Informed consent for participation in the study was obtained from all patients.
Clinical assessment and outcomes
The following parameters were recorded: primary tumor size, tumor grade, tumor, node, metastasis (TNM) staging, vascular invasion, alpha-fetoprotein (AFP) level, and hepatitis B DNA quantification one month after surgery. According to the use of sirolimus in the immunosuppressive regimen after transplantation by each patient, the participants were divided into a sirolimus group (n=35) and a non-sirolimus group (n=11).
Experimental procedures
Sample collection
The follow-up date began in December 2012, and the end date was August 2019. Follow-up was mainly performed at outpatient visits and partially via telephone communication and was terminated when the patient died. There were no patients lost to follow-up. A Cox survival curve was used to analyze patients’ survival after transplantation, and logistic regression analysis was used to analyze patients’ prognostic factors in both groups. All HCC tumor specimens were frozen in liquid nitrogen immediately after surgical resection in the operating room and stored at –80 °C. Samples were obtained from the tumor, and paraffin sections were then prepared for immunohistochemical and histological examinations. FFPE tissue specimens and matched blood samples were collected after liver transplantation for HCC, and targeted next-generation sequencing was performed by using solid tumor-associated genes (Genetron Health; Beijing, China). These included recurrent mutations in TP53, Wnt-signaling components CTNNB1 and AXIN1, and chromatin regulators like ARID1A and ARID2, and several important genes related to the mTOR pathway, including TSC1/2, mTOR, MAPK1, PTEN, PIK3CG, and EIF4E2.
Immunohistochemistry
(I) Two consecutive sections (5 µm thick) of the same paraffin-embedded tissue were obtained. (II) The paraffin sections were deparaffinized with xylene, hydrated with graded alcohol, and washed with PBS three times for 3 minutes each. (III) Antigen retrieval solution was performed under high pressure and washed with PBS three times for 3 minutes each. (IV) 3% hydrogen peroxide (50 µL) was added to each section, and the sections were incubated at room temperature for 10 minutes to block endogenous peroxidase activity and then washed with PBS three times for 3 minutes each. (V) The PBS solution was drained, and 50 µL of TSC primary antibody (RRID: AB_217328) (antibody working concentration of 1:100) was added to each section; the sections were incubated for 2 hours at room temperature, followed by a PBS washing three times for 5 minutes each. (VI) The PBS solution was aspirated, and 50 µL of polymer enhancer (reagent A) was added to each section; the sections were incubated at room temperature for 20 minutes and washed with PBS three times for 3 minutes each time. (VII) The PBS solution was drained, and 50 µL of enzyme-labeled anti-rat/rabbit polymer (reagent B) was added dropwise to each section; the sections were incubated at room temperature for 30 minutes, followed by a wash with PBS three times for 3 minutes each time. (VIII) The PBS solution was drained, and 50 µL of freshly prepared DAB solution was added to each section. After microscopic confirmation of staining intensity, the reaction was stopped, and the sections were rinsed with tap water. (IX) The sections were counterstained with hesperidin, and 0.1% HCl solution was added to the sections and washed off with tap water for 15 minutes. (X) The sections were dehydrated with a graded alcohol series and cleared with xylene and then sealed with neutral mounting media.
Gene sequencing
Purified DNA from formalin-fixed paraffin-embedded (FFPE) tissue samples from the 46 HCC patients who underwent liver transplantation were examined. The QIAampDNA FFPE kit (Qiagen, Hilden, Germany) was employed, following the manufacturer’s instructions, and concentrations of the purified DNA were determined using the Qubit ds DNA assay (Life Technologies Gaithersburg, USA). Samples with ≥2.5 ng DNA/mL were diluted to this concentration, and genomic DNA from those samples was extracted and captured using a panel of genes related to HCC development. High-throughput sequencing was performed on an Illumina HiSeq 4000 platform (Genetron Health, Beijing, China), and sequencing reads were mapped to a human reference genome (hg19) using the Burrows-Wheeler Aligner (BWA). Duplicate removal, local realignment, and base quality recalibration were performed using PICARD (http://broadinstitute.github.io/picard/) and the Genome Analysis Toolkit (GATK) (22). Somatic single nucleotide variations (SNVs) and small indels were detected using Mutect and Strelka (23).
Statistical analysis
Statistical analysis was performed using SPSS 22.0 software. The results were expressed as the frequency and percentage. Fisher’s exact test was used in the univariate analysis for comparing categorical variables between the two groups, and multivariate analysis of the prognostic factors for the overall survival curve was performed using the Cox proportional hazards model and survival analyses. The cumulative survival time was calculated using the Kaplan-Meier method, and P<0.05 was considered the standard of statistical significance.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University ([2015]2-167). And informed consent was taken from all the patients.
Results
Clinical outcomes
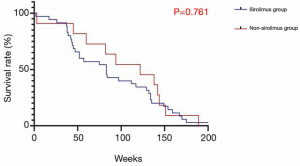
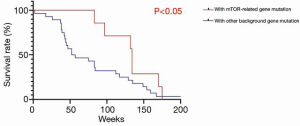
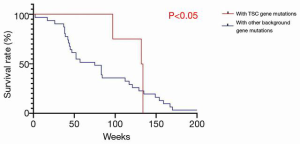
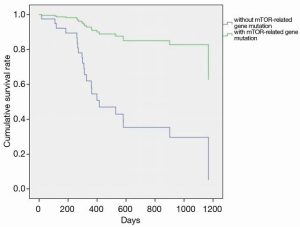
The characteristics of the 46 patients who underwent liver transplantation for HCC are shown in Table 1. The average age of the sample population was 51±21 years; there were 45 males and 1 female; 65.2% (n=30) of the patients had primary tumors larger than 5 cm in diameter; 67% (N=31) of the patients had tumors macrovascular invasion; 78.2% (n=36) of the tumors were TNM stage III and above. The overall characteristics of the sample population were older patients with advanced HCC (Milan criteria). While there was no significant difference in survival rates between the two groups (P=0.761) (Figure 1), in the sirolimus group, patients with mTOR-related gene mutations (Figure 2) had significantly better survival rates than those with other types of gene mutations (P<0.05). Consistent with this, the survival rate of patients with TSC gene mutations was also significantly better than that of patients without TSC mutations (P<0.05) (Figure 3). A Cox model was used to analyze the survival rates of patients in the sirolimus group (n=35) and the non-sirolimus group (n=11) (P<0.05) (Figure 4). We found that patients with mutations in mTOR pathway-related genes had better survival rates than patients without mutations in mTOR pathway-related genes. This mutation is associated with decreased expression or inactivity of TSC-encoded protein complexes. As it is a negative regulator of the mTOR signaling pathway, its decreased expression or inactivity leads to the abnormal activation of the mTOR signaling pathway (24). This means that tumor cell growth and proliferation are more active but more sensitive to mTOR inhibitors, such as sirolimus. This is the reason that the prognosis of HCC patients with TSC1/2 mutations is better than that of those without TSC1/2 mutations when undergoing sirolimus treatment. Taken together, we confirmed that TSC1/2 mutations lead to unique pathological types of HCC. Although these tumor types are more biologically aggressive, patients with these genotypes have a better prognosis while using sirolimus after liver transplantation for HCC.
Full table
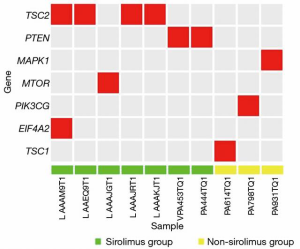
Genetic test results
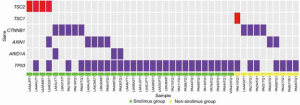
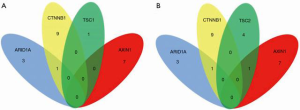
Several key genes (TSC1/2, mTOR, MAPK1, PTEN, PIK3CG, and EIF4E2) in the mTOR pathway were tested in FFPE tissue samples from the 46 patients. The results show that 10 patients carried the relevant mutations (Figure 5), including seven in the sirolimus group and three patients in the non-sirolimus group. Five patients carried a TSC1/2 mutant gene, almost accounting for 50% of all mTOR-related gene mutations. Also, we examined the CTNNB1, AXIN1, ARID1A, and TP53 genes, which play important roles in human HCC and have a high frequency of mutations (Figure 6). The gene with the highest mutation frequency was TP53, occurring in 82.6% (n=38) of the 46 samples, followed by CTNNB1, occurring in 19.6% (n=9), AXIN1 (n=7, 15.2%), and TSC1/2 (n=5, 10.9%). The gene with the lowest mutation frequency was ARID1A (n=3, 6.5%), and it can be seen from the Venn diagram (Figure 7) that TSC1/2 and other genes do not significantly cross.
Comparison between TSC1/TSC2 mutations and other pathological background mutations
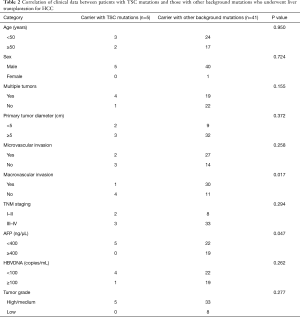
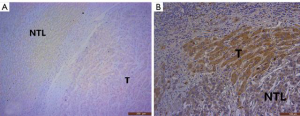
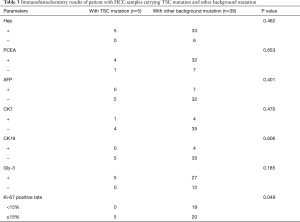
Comparing the clinical data of patients with TSC mutations versus other background mutations, we found that the incidence of major vascular invasion (P=0.017) and serum AFP concentration (P=0.047) were significantly different between those with TSC1/2 mutations (n=5) and patients with other background mutations (n=41). However, there were no statistically significant differences in tumor diameter (P=0.372), TNM stage (P=0.294), and degree of tumor differentiation (P=0.277), which are related to tumor malignancy and prognosis (Table 2). Also, we performed immunohistochemistry on sections obtained from the 46 patients and found that in sections with TSC1/2 mutations, expression intensity in tumor tissue was lower than that in nontumor tissue. In contrast, in the samples obtained from patients without TSC1/2 mutation, there was no significant difference in the expression intensity between tumor tissue and nontumor liver tissue (Figure 8A,B). Moreover, we compared the differences in multiple immunohistochemical indicators between patients with TSC1/2 mutations and those with other background mutations (Table 3). The results showed that in terms of the expression of proliferating cell nuclear antigen Ki-67, Ki-67 positive rates were ≥15% in the samples from patients carrying TSC1/2 mutations (n=5), and there was a significant difference between the two groups (P=0.038).
Full table
Full table
Discussion
Much attention has been paid to the application of mTOR pathway inhibitors, such as sirolimus, in liver transplantation patients because of the strong immunosuppressive effects and anti-cancer effects of these inhibitors (25-27). Many studies have shown that sirolimus can reduce the tumor recurrence rate and improve the survival rate of patients undergoing liver transplantation for HCC (20,28,29). However, in the 46 patients with HCC in this study, there was no significant difference in survival between the sirolimus and non-sirolimus groups. Our results echo those of a prospective Phase III multi-center randomized controlled trial, which also showed no difference in the five-year disease-free survival rate of two groups of patients who received or did not receive treatment with sirolimus (30). However, our further analysis of their gene sequencing results showed that patients with mTOR-related gene mutations in the sirolimus group had significantly better survival rates than patients without mTOR-related gene mutations. The key role of the mTOR pathway in cell growth and survival makes it a reasonable target for anti-tumor strategies, as evidenced by various clinical data types (31). To study the efficacy of mTOR signaling pathway mutation in the treatment of HCC after liver transplantation more accurately, we evaluated the gene sequencing results of the 46 patients and found that there were 10 samples with mutations in mTOR pathway-related genes (including TSC1/2, PTEN, MAPK1, PI3KCG, EIF4A2, and mTOR), of which 50% were TSC1/2 mutations. These TSC1/2 mutations are deleterious mutations for the TSC protein and often decrease TSC expression or loss of TSC activity (19). Our immunohistochemical results confirmed this finding. Based on the exclusive mode between the high mutation frequency of TSC (10.9%) and the key mutated genes in HCC (TP53, CTNNB1, AXIN1, and ARID1A) in existing studies (Figure 6), we have reason to believe that TSC mutations are major driving factors in the development of HCC and are no less important than known HCC driving factors. As TSC is a key negative regulator of the mTOR pathway, frequent TSC mutations and inactivation are likely to be important reasons for the mTOR pathway’s excessive activation (32,33). According to existing studies, a reduction in TSC expression is associated with advanced tumor progression, vascular invasion, and poor prognosis in HCC patients (34,35). In our study, inpatients taking sirolimus, the survival rate of patients with TSC1/2 mutations was significantly better than that of patients without TSC mutations (Figure 3). This suggests that patients with these mutations are likely to gain potential benefits from the use of sirolimus after liver transplantation for HCC. This also provides a possible theoretical basis for treating some patients with TSC1/2 mutations but who are in advanced stages of HCC. TSC1/2 mutations can be used as recognized biomarkers to predict the efficacy of mTORC1 inhibitors (36). Our study supports the rationale for using mTOR inhibitors (sirolimus) in patients undergoing liver transplantation for HCC who have mTOR-related gene mutations (especially TSC mutations).
Interestingly, the research of Janku (37) showed a high frequency of mTOR-related mutations in HCC patients. Hao (33) confirmed the existence of over-activation of the mTOR pathway during the occurrence of HCC, and in their study TSC2 was often lost in the samples. These findings provide more support for the hypothesis that “TSC has an inactive mutation—mTOR through excessive activation—HCC development” and highlights the unique position of the TSC1/2 mutation in this process. Therefore, for liver cancer patients with TSC1/2 gene mutation detected before surgery, whether preoperative mTOR inhibitor (sirolimus) adjuvant therapy can improve liver transplantation prognosis is also worth exploring.
Conclusions
In summary, we suggest performing genetic tests in HCC patients before liver transplantation. The results will help develop a personalized treatment, predict the prognosis, and extend the surgical indications for some HCC patients. This study’s limitations include its small sample size, single-center conduct, and retrospective nature, and we are currently conducting a prospective study to address the latter. However, the sample size has been quite large in the genetic testing of single-center liver transplantation of HCC. We conclude that more HCC patients with over-standard but with TSC1/2 mutations have achieved good prognosis after liver transplantation and treatment with sirolimus from the limited data available.
Acknowledgments
The authors thank Mengchen Shi for her critical editing of this manuscript.
Funding: This work was supported by the National 13th Five-Year Science and Technology Plan Major Projects of China (2017ZX10203205), National Key R&D Plan (2017YFA0104304), National Natural Science Foundation of China (81770648, 81972286), Guangdong Natural Science Foundation (2018A030313259, 2015A030312013), Science and Technology Program of Guangdong Province (2017B020209004, 20169013, 2020B1212060019), Science and Technology Program of Guangzhou City (201508020262), Guangdong Basic and Applied Basic Research Foundation (2019A1515110654, 2020A1515010574), the Fundamental Research Funds for the Central Universities (20ykpy38), and China Postdoctoral Science Foundation (2019TQ0369, 2020M672987).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-378
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-378
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-378). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University ([2015]2-167). And informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.、
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418-9. [Crossref] [PubMed]
- Duffy JP, Vardanian A, Benjamin E, et al. Liver transplantation criteria for hepatocellular carcinoma should be expanded: a 22-year experience with 467 patients at UCLA. Ann Surg 2007;246:502-9; discussion 509-11. [Crossref] [PubMed]
- Sotiropoulos GC, Molmenti EP, Losch C, et al. Meta-analysis of tumor recurrence after liver transplantation for hepatocellular carcinoma based on 1,198 cases. Eur J Med Res 2007;12:527-34. [PubMed]
- Santopaolo F, Lenci I, Milana M, et al. Liver transplantation for hepatocellular carcinoma: Where do we stand? World J Gastroenterol 2019;25:2591-602. [Crossref] [PubMed]
- Lin HS, Wan RH, Gao LH, et al. Adjuvant chemotherapy after liver transplantation for hepatocellular carcinoma: a systematic review and a meta-analysis. Hepatobiliary Pancreat Dis Int 2015;14:236-45. [Crossref] [PubMed]
- Söderdahl G, Bäckman L, Isoniemi H, et al. A prospective, randomized, multi-centre trial of systemic adjuvant chemotherapy versus no additional treatment in liver transplantation for hepatocellular carcinoma. Transpl Int 2006;19:288-94. [Crossref] [PubMed]
- Bassanello M, Vitale A, Ciarleglio FA, et al. Adjuvant chemotherapy for transplanted hepatocellular carcinoma patients: impact on survival or HCV recurrence timing. Transplant Proc 2003;35:2991-4. [Crossref] [PubMed]
- Field KM, Michael M, Part II. Liver function in oncology: towards safer chemotherapy use. Lancet Oncol 2008;9:1181-90. [Crossref] [PubMed]
- Wang G, Zhang Z. Evaluation and prospect of chemical therapeutic drug of primary hepatocellular carcinoma . Drug Eval 2009;6:169-72.
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90. [Crossref] [PubMed]
- Cheng AL, Kang YK, Chen Z, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol 2009;10:25-34. [Crossref] [PubMed]
- Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;389:56-66. [Crossref] [PubMed]
- Villanueva A, Chiang DY, Newell P, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology 2008;135:1972-83, 1983.e1-11.
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell 2007;12:9-22. [Crossref] [PubMed]
- Wang Z, Zhou J, Fan J, et al. Sirolimus inhibits the growth and metastatic progression of hepatocellular carcinoma. J Cancer Res Clin Oncol 2009;135:715-22. [Crossref] [PubMed]
- Guba M, von Breitenbuch P, Steinbauer M, et al. Rapamycin inhibits primary and metastatic tumor growth by antiangiogenesis: involvement of vascular endothelial growth factor. Nat Med 2002;8:128-35. [Crossref] [PubMed]
- Zimmerman MA, Trotter JF, Wachs M, et al. Sirolimus-based immunosuppression following liver transplantation for hepatocellular carcinoma. Liver Transpl 2008;14:633-8. [Crossref] [PubMed]
- Toso C, Merani S, Bigam DL, et al. Sirolimus-based immunosuppression is associated with increased survival after liver transplantation for hepatocellular carcinoma. Hepatology 2010;51:1237-43. [Crossref] [PubMed]
- Duvoux C, Toso C. mTOR inhibitor therapy: Does it prevent HCC recurrence after liver transplantation? Transplant Rev (Orlando) 2015;29:168-74. [Crossref] [PubMed]
- Menon KV, Hakeem AR, Heaton ND. Meta-analysis: recurrence and survival following the use of sirolimus in liver transplantation for hepatocellular carcinoma. Aliment Pharmacol Ther 2013;37:411-9. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- Ferrín G, Guerrero M, Amado V, et al. Activation of mTOR Signaling Pathway in Hepatocellular Carcinoma. Int J Mol Sci 2020;21:1266. [Crossref] [PubMed]
- Ho DW, Chan LK, Chiu YT, et al. TSC1/2 mutations define a molecular subset of HCC with aggressive behaviour and treatment implication. Gut 2017;66:1496-506. [Crossref] [PubMed]
- Geissler EK, Schnitzbauer AA, Zulke C, et al. Sirolimus Use in Liver Transplant Recipients With Hepatocellular Carcinoma: A Randomized, Multicenter, Open-Label Phase 3 Trial. Transplantation 2016;100:116-25. [Crossref] [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28:1811-7. [Crossref] [PubMed]
- Soll C, Clavien PA. Inhibition of mammalian target of rapamycin: two goals with one shot? J Hepatol 2011;54:182-3. [Crossref] [PubMed]
- Navarro-Villarán E, de la Cruz-Ojeda P, Contreras L, et al. Molecular Pathways Leading to Induction of Cell Death and Anti-Proliferative Properties by Tacrolimus and mTOR Inhibitors in Liver Cancer Cells. Cell Physiol Biochem 2020;54:457-73. [Crossref] [PubMed]
- Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front Oncol 2014;4:64. [Crossref] [PubMed]
- Toso C, Meeberg GA, Bigam DL, et al. De novo sirolimus-based immunosuppression after liver transplantation for hepatocellular carcinoma: long-term outcomes and side effects. Transplantation 2007;83:1162-8. [Crossref] [PubMed]
- Huynh H, Hao HX, Chan SL, et al. Loss of Tuberous Sclerosis Complex 2 (TSC2) Is Frequent in Hepatocellular Carcinoma and Predicts Response to mTORC1 Inhibitor Everolimus. Mol Cancer Ther 2015;14:1224-35. [Crossref] [PubMed]
- Huang KT, Huang YH, Li P, et al. Correlation between tuberous sclerosis complex 2 and glycogen synthase kinase 3 beta levels, and outcomes of patients with hepatocellular carcinoma treated by hepatectomy. Hepatol Res 2014;44:1142-50. [Crossref] [PubMed]
- Kenerson HL, Yeh MM, Kazami M, et al. Akt and mTORC1 have different roles during liver tumorigenesis in mice. Gastroenterology 2013;144:1055-65. [Crossref] [PubMed]
- Menon S, Yecies JL, Zhang HH, et al. Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Sci Signal 2012;5:ra24. [Crossref] [PubMed]
- Janku F, Kaseb AO, Tsimberidou AM, et al. Identification of novel therapeutic targets in the PI3K/AKT/mTOR pathway in hepatocellular carcinoma using targeted next generation sequencing. Oncotarget 2014;5:3012-22. [Crossref] [PubMed]
(English Language Editors: B. Draper and J. Chapnick)


