
Adaptive immunity profiling associated with histological subtypes and postoperative survival in gastric cancer
Introduction
Gastric cancer (GC) is one of the most common human malignancies and the prognosis of advanced disease remains dismal (1). The substantial morbidity and mortality rates of GC has made it the fifth most commonly diagnosed cancer and the third leading cause for cancer-related death, with 1,000,000 newly diagnosed cases and 783,000 deaths annually (2). The pathogenesis of human GC is thought to be greatly affected by genetic and epigenetic mutations, including the overexpression of oncogenes and the downregulation of tumor suppressor genes (3). Recently, immune checkpoint inhibitors, for example anti-programmed cell death-1 (PD-1) or programmed cell death ligand-1 (PD-L1) monoclonal antibodies, have objectively improved the overall survival (OS) of patients with various types of carcinomas including advanced GC. Recently, there were two anti-PD-1 inhibitors approved for treatment of advanced GC in Japan: nivolumab (as third- or later-line treatment for AGC) and pembrolizumab. However, only a subset of patients showed a clinical benefit, indicating the heterogeneity of GC etiology, and revealing the importance of accurate patient selection and specific treatment to enhance sensitivity for PD-1/PD-L1 blockade (4).
In tumor immunity, adaptive immunity is defined by the presence of lymphocytes such as T cells or B cells. During the process of tumorigenesis, CD8+ cytotoxic T cells act as effector cells that directly destroy tumor cells, CD4+ helper T cells regulate CD8+ T- and B-cell function, and B cells present antigens and produce antibodies (5). Although several studies have provided evidence supporting the evolution of tumor immune escape, the role of the adaptive immune response in influencing the progression and recurrence of tumors has long been controversial (6). Some studies have suggested that the adaptive immune response participates in the damage of H. pylori infection and partly facilitates tumor development, and that tumor-associated monocytes/macrophages suppress the human NK-cell (natural killer cell) function in GC (7,8). In contrast, studies focused on the specific molecules have indicated that a stress-induced glucocorticoid surge and Tsc22d3 upregulation affect therapy-induced anti-cancer immunosurveillance (9), and that HSP-gp96 is capable of stimulating T cells and DC cells (dendritic cell) and inducing cytokine secretion in GC (10). Previous study has indicated that the polymorphisms in inflammatory response-related genes were involved in the pathogenesis of gastric cancer (11). The gene expression regulation of adaptive immunity also showed a prognostic role in GC (12). Multiple studies have demonstrated adaptive immune cell and molecule variations in GC (8,13,14); however, a systematic evaluation of adaptive immunity-related gene expression in the inner tumor tissues is absent. In this study, we assessed a comprehensive expression profile of molecules involved in adaptive immunity in 412 primary GC surgical excision tissues from The Cancer Genome Atlas (TCGA) project and investigated their correlations to clinical manifestations. Moreover, we studied the different expressing genes between the multiple histological types and uncovered their prognostic roles.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jgo-21-79).
Methods
Patients
Information regarding patients with GC and their corresponding clinical data were collected from TCGA database (cbioportal.org) (15). All transcriptome data were from specimens taken from the primary tumor site. Informed consent was verified according to TCGA standards. Among the studied cohort, 412 subjects were enrolled. There were seven pathological types, including diffuse type stomach adenocarcinoma (DSTAD, n=69), intestinal type stomach adenocarcinoma (ISTAD, n=72), mucinous stomach adenocarcinoma (MSTAD, n=20), papillary stomach adenocarcinoma (PSTAD, n=7), signet ring cell carcinoma (SSRCC, n=12) of stomach adenocarcinoma (STAD, n=156), and tubular stomach adenocarcinoma (TSTAD, n=76). All the data involving human are from the public database (cbioportal.org) and the ethical statements is obtained before. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013) (16).
RNA sequencing data and clinical features
We filtered the data according to whether the clinical features (including survival data) and mRNA z-score data were completed. Patients underwent postoperative pathological diagnosis and had detailed clinical records, including age, sex, grade, aneuploidy score, and fraction genome altered, were enrolled in the current study. According to the histological records, all of the samples were diagnosed as GC with different histological subtypes. The patients received surgical treatment and RNA sequencing was performed on the excised tissues. Collectively, there were 412 samples enrolled in the final assessment. All utilized public omics data sets were previously generated by reported studies and the prior ethical approvals have been obtained.
Bioinformatics
Adaptive immunity-associated genes analyzed in the current study were collected from the Uniport-keyword database (KW-1064, uniport.org/keywords) (17). Excluding those with missing information, 158 genes reported to participate in adaptive immunity were evaluated, and their expressional data (RNA-Seq V2 RSEM) were collected from TCGA database. A cluster analysis of gene expression in GC patients was performed to divide samples into different groups based on gene expression profiles. Cases with similar expressional patterns were identified from the entire samples. The transcriptional levels of genes were shown as mRNA z-scores and clustered using the hierarchical clustering algorithm via a program developed by Stanford (Michael Eisen, http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm#ctv) (18). The cluster heat map and the genes expressional patterns according to tumor stage were generated using the Java Treeview program (http://jtreeview.sourceforge.net/) (19). The VENNY diagram was generated by an online tool: BioVen (BioinfoGP, https://bioinfogp.cnb.csic.es/tools/venny/) (20).
Prognostic relevance analyses
The prognostic roles of the adaptive immunity-associated genes were evaluated via comparing the survivals of different cohorts. The OS, progression-free survival (PFS), disease-free survival (DFS), and disease-specific survival (DSS) were calculated using GraphPad Prism software (GraphPad Software, Inc., California, US; Version 8). Comparison of survivals between different clusters revealed the relevance of prognosis and gene expressional profiles. Additionally, the OS corresponding to each subgroup with low or high expression levels of single gene were analyzed using GraphPad Prism 8.
Statistical analysis
Survival curves corresponding to different subgroups were plotted and compared using the log-rank (Mantel-Cox) test in GraphPad Prism 8. Clinical characteristics and variables related to each cluster were compared and assessed by Fisher’s exact test and Pearson correlation analysis. Differentially Expressed Genes between subgroups were identified using ANOVA (analysis of variance). Correlations between variables or genes expressions were determined according to regression analyses. All tests were performed with SPSS 24.0 (IBM, Inc., New York, US). P values less than 0.05 were considered statistically significant.
Results
Adaptive immunity-associated genes were significantly varied between each GC histological type
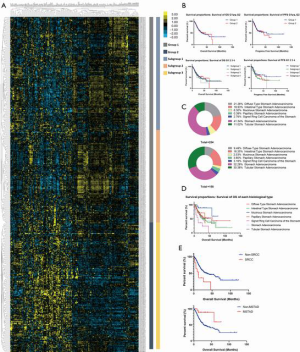
According to hierarchical clustering of the matrix, which consisted of expressional information for the adaptive immunity-associated genes of each patient with GC, two primary groups and four subdivided groups were identified from the entire cohort (Figure 1A). OS and PFS comparison between each primary or subdivided cluster showed no differences (Figure 1B, Table 1). Our analysis of the proportion of histological types showed that the group 1 was dominated by STAD (41.34%) and DSTAD (21.26%), while group 2 was dominated by STAD (32.28%) and TSTAD (30.38%) (Figure 1C). We compared the outcomes of each histological type and detected a significant difference in the OS (Figure 1D, P=0.014), of which SRCC showed the worst OS and MSTAD showed the longest OS (Table 2). Additional assessments confirmed that patients with SRCC type had a worse OS compared to those with non-SRCC types (12.43 vs. 40.00 months, P=0.004), and patients with MSTAD type showed a favorable OS compared to those with other types (undefined vs. 28.57 months, P=0.010) (Figure 1E, Table 2).

Full table

Full table
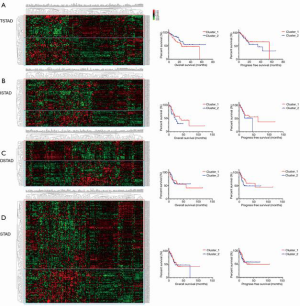
We additionally divided the patients into different cohorts according to histological types, and identified subgroups in the TSTAD, ISTAD, DSTAD, and STAD types. Patients with common histological types were grouped into two clusters, however there were no differences detected in the OS or PFS among each histological type (Figure 2 and Table S1).
Subgroups with characteristic adaptive immunity-associated gene expression were correlated to the tumor stage and grade
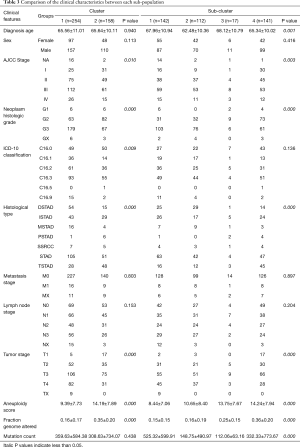
To investigate the correlation between adaptive immunity and clinical performance, we compared the clinical features between each identified group or subgroup. For groups 1 and 2, the AJCC stage, grade, histological type, tumor stage, aneuploidy score, and fraction genome altered were significantly different (P<0.05, Table 3). Among subdivided groups 1 to 4, age, AJCC stage, grade, histological type, tumor stage, aneuploidy score, fraction genome altered, and mutation count were significantly different (P<0.05, Table 3). Compare to cluster_2, the cluster_1 showed more cases with advanced AJCC stage (54.33% vs. 48.10% of stage III&IV), higher grade (70.47% vs. 42.41%), STAD type (41.34% vs. 32.28%), and advanced tumor stage (74.02% vs. 67.09% of T3&T4), on the other side, patients in cluster_1 showed more C16.3 type (9.07% vs. 31.65%), low aneuploidy score (9.39±7.73 vs. 14.19±7.89) and less fraction genome altered (0.16±0.17 vs. 0.35±0.20). Of these discrepant features, AJCC stage, tumor stage, and grade were the commonly changed elements among both primary and secondary classifications.
Full table
Adaptive immunity-associated genes were significantly varied between histological types of GC
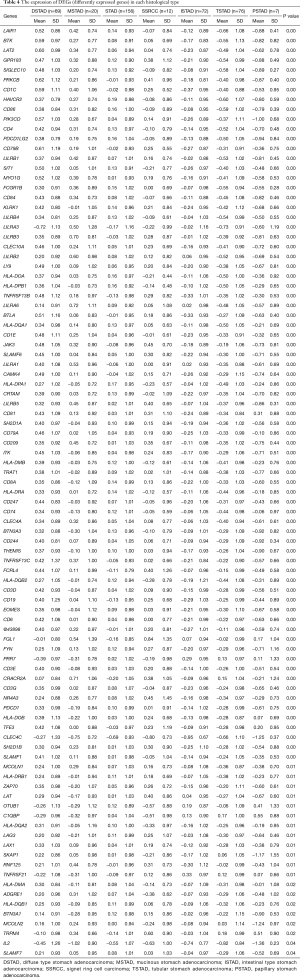
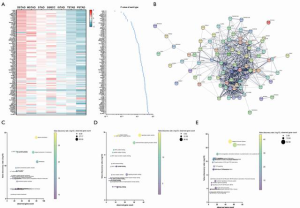
We detected that patients with different histological types showed discrepant prognoses and those types in each identified subgroup were differently distributed (Figure 1). To assess the histological type-correlated factors, we compared the gene expression of adaptive immunity between each of the seven types. In total, there were 95 differently expressed genes (DEGs) detected, which were represented by LAIR1, BTK, and LAT2 (Table 4). According to the expressional profiling, patients with DSTAD, STAD, MSTAD, and SSRCC types showed a similar adaptive immunity form, otherwise, patients with TSTAD, ISTAD and PSTAD showed a different adaptive immunity form (Figure 3A). The functional network indicated that the DEGs were involved in multiple biological events (Figure 3B). GO (Gene Ontology) analysis of cellular components showed that these DEGs were dominantly distributed in the plasma membrane and MHC class II protein complex, such as CD74 and HLA-DOA (Figure 3C). GO analysis of molecular function indicated that most DEGs were principally involved in antigen binding and signaling receptor activity, including LAG3 and LILRA1 (Figure 3D). Reactome pathway analysis revealed that these genes managed the immunoregulatory interaction between lymphoid and a non-lymphoid cell (CRTAM and LILRB3, for instance) and participated in PD-1 signaling (CD4 and CD3D, for example) (Figure 3E).
Full table
Adaptive immunity-associated genes were capable of distinguishing between different histological types of GC
We found that patients with SRCC and MSTAD histological types exhibited the worst and the best prognosis, respectively. To elucidate the relevance of adaptive immunity and the two types of GC, we assessed the DEGs between patients with each type and the other types. For the MSTAD type, there were 11 genes, represented by LILRA6 and LILRA3, which were significantly overexpressed (Figure 4A,B). According to the regression analysis, the model combining seven genes showed an excellent ability to recognize the MSTAD type (Table S2). The area under the ROC (receiver operating characteristic curve) curve was 0.91 (Figure 4C), and the model to differentiate whether the histological type was MSTAD was based on the predicted value (Figure 4D). Similarly, we found that six genes, represented by FGL1 and SKY, were differently expressed between SRCC and the other types (Figure 4E,F). A regression analysis showed that the model combining six genes demonstrated an ability to distinguish the SRCC type from the other types (AUC =0.91, Figure 4G). According to this model, patients with SRCC or non-SRCC were well defined (Figure 4H).
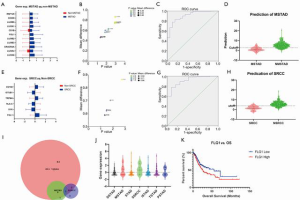
We detected that the 95 adaptive immunity-associated genes were significantly varied between each histological type. To study the commonly changed genes of the different types and the overlapping DEGs among all types, MSTAD and SRCC were assessed. Only FGL1 was identified in the overlaps (Figure 4I). Moreover, FGL1 was highly expressed in MSTAD and SRCC (Figure 4J). Analysis of the correlation with prognosis showed that the high expression of FGL1 was strongly associated with a worse OS; the median survival time was 55.43 months in the low-expression group and 25.71 months in the high expression group (P=0.015, Figure 4K).
Discussion
Aberrant expression of immunity-associated markers is significantly associated with solid tumor progression and postoperative prognosis (21). Adaptive immunity refers to the protection of a host organism from a pathogen or toxin mediated by B and T cells, and is characterized by immunological memory. Unlike other immune responses, adaptive immunity is highly dependent on a given antigen or abnormal internal environment. Numerous studies have demonstrated the aberrant immunity status in human cancers and suggested the prognostic role of genes involved in immune regulation. However, there are few studies on the expressional profiles of adaptive immunity-related genes and their implication on clinical features in GC. In the current study, we identified different sub-populations according to the gene expressional pattern and investigated their correlation with multiple clinical manifestations.
Regardless of the specific histological type, the enrolled GC patients were clustered into different groups according to the expressional profiles of adaptive immunity-related genes using dichotomy method or trichotomy method (Figure 1). OS and PFS comparisons revealed no differences between each group. Even among patients with a common histological type, no differences were detected between subgroups showing similar expression patterns. This data demonstrated that the synthetic genes’ expression did not directly influence GC prognosis. Assessment of the histological proportions of two primary groups showed that group 1 was constituted mainly of STAD and ISTAD, while group 2 primarily comprised STAD and TSTAD. This result suggested that the expression pattern of adaptive immunity-related genes was closely connected to the difference in histological type. In addition to histological type, the AJCC stage, grade, and tumor stage were notably different between each sub-population (Table 3). These results demonstrated that changes in adaptive-immune status were significantly correlated with the clinical characteristics of GC.
Our comparison of the DEGs pattern of different histological types revealed that patients with DSTAD, STAD, MSTAD, and SSRCC types showed a similar expressional profile, while patients with TSTAD, ISTAD, and PSTAD exhibited an opposite expressional profile (Figure 3). The functional network and enrichment analysis indicated that these DEGs were dominantly distributed in the plasma membrane and MHC class II protein complex, and were principally involved in antigen binding and signaling receptor activity, as well as immunoregulatory interaction between lymphoid and non-lymphoid cells. Moreover, these DEGs participated in PD-1 signaling. Our data demonstrated the discrepant adaptive-immune molecular expression and signaling among different histological types.
Although previous studies reported that patients with different histological types had varying prognoses; in the current study, we evaluated the postoperative survival in each histological type. Compared to the other types, patients with SRCC and MSTAD exhibited the worst and best prognoses, respectively. Assessment of DEGs showed that 11 genes were significantly overexpressed in the MSTAD and six genes exhibited opposite expression between the SRCC type and other types (Figure 4). To distinguish the specific histological types, we performed regression analysis based on gene expression. The model combining seven genes was acceptable to recognize the MSTAD type (AUC =0.91) and the model combining six genes showed an ability to distinguish the SRCC type from other types (AUC =0.91). According to this model, patients with SRCC or non-SRCC were well defined (Figure 4). Additionally, we discovered that FGL1 was differently expressed between each histological type, and that the elevated expression was correlated with a poor OS in all patients. FGL1 is the coding gene of fibrinogen-like protein 1, which is specifically secreted by the liver and reported to be a major LAG-3 functional ligand independent from MHC-II (22). A previous study demonstrated that suppression of the FGL1-LAG-3 interaction could stimulate tumor immunity (22). This result might partly support our finding. FGL1 is highly expressed by human cancer cells, yet few studies have focused on FGL1 expression in GC tissues or the regulatory effect of FGL1 in GC (23). A previous study revealed that FGL1 expression was upregulated in GC tissues, and that high FGL1 expression levels strongly indicated a shorter OS time of GC patients (P=0.005), and significantly decreased the expression of N-cadherin and vimentin levels in GC cells (P<0.01) (24). This result is consistent with our data and indicates the potential regulatory mechanism of FGL1 in GC. Designing the molecule inhibitors or antibody targeted the specific genes (or proteins) playing a prognostic role is a feasible strategy for the precisely therapy.
In conclusion, our study demonstrated the expressional profile of the 158 adaptive immunity-related genes in 412 patients, and detected the specific gene expression patterns among multiple histological types. The comparison of clinical features between each immune subgroup revealed that the expressional profile was unable to predict prognosis, but was capable of distinguishing the different histological types. The expression of an individual gene, like FGL1, was significantly associated with GC prognosis. These results are expected to promote the tumor immunological mechanisms in GC and should be verified by further research.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jgo-21-79
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-21-79). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Moehler M, Dvorkin M, Boku N, et al. Phase III Trial of Avelumab Maintenance After First-Line Induction Chemotherapy Versus Continuation of Chemotherapy in Patients With Gastric Cancers: Results From JAVELIN Gastric 100. J Clin Oncol 2021;39:966-77. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Kawazoe A, Shitara K, Boku N, et al. Current status of immunotherapy for advanced gastric cancer. Jpn J Clin Oncol 2021;51:20-7. [Crossref] [PubMed]
- Stanton SE, Ramos E, Disis ML. 71 - Immunologic Approaches to Breast Cancer Therapy. In: Bland KI, Copeland EM, Klimberg VS, et al. editors. The Breast (Fifth Edition): Elsevier, 2018:924-933.e4.
- Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960-4. [Crossref] [PubMed]
- Yolanda LV, Sergio PD, Hugo ES, et al. Gastric cancer progression associated with local humoral immune responses. BMC Cancer 2015;15:924. [Crossref] [PubMed]
- Peng LS, Zhang JY, Teng YS, et al. Tumor-Associated Monocytes/Macrophages Impair NK-Cell Function via TGFβ1 in Human Gastric Cancer. Cancer Immunol Res 2017;5:248-56. [Crossref] [PubMed]
- Yang H, Xia L, Chen J, et al. Stress-glucocorticoid-TSC22D3 axis compromises therapy-induced antitumor immunity. Nat Med 2019;25:1428-41. [Crossref] [PubMed]
- Lu WW, Zhang H, Li YM, et al. Gastric cancer-derived heat shock protein-gp96 peptide complex enhances dendritic cell activation. World J Gastroenterol 2017;23:4390-8. [Crossref] [PubMed]
- Furuya TK, Jacob CE, Tomitão MTP, et al. Association between Polymorphisms in Inflammatory Response-Related Genes and the Susceptibility, Progression and Prognosis of the Diffuse Histological Subtype of Gastric Cancer. Genes (Basel) 2018;9:631. [Crossref] [PubMed]
- Liu F, Bu Z, Zhao F, et al. Increased T-helper 17 cell differentiation mediated by exosome-mediated microRNA-451 redistribution in gastric cancer infiltrated T cells. Cancer Sci 2018;109:65-73. [Crossref] [PubMed]
- Zhong C, Song Z, Li M. Gastric cancer patients display a distinctive population of IFNg(+)IL10(+) double positive CD8 T cells, which persists longer during prolonged activation. Exp Cell Res 2019;382:111487 [Crossref] [PubMed]
- Nagase H, Takeoka T, Urakawa S, et al. ICOS(+) Foxp3(+) TILs in gastric cancer are prognostic markers and effector regulatory T cells associated with Helicobacter pylori. Int J Cancer 2017;140:686-95. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- World Medical Association Declaration of Helsinki. ethical principles for medical research involving human subjects. JAMA 2013;310:2191-4. [Crossref] [PubMed]
- Ata SK, Fang Y, Wu M, et al. Disease Gene Classification with Metagraph Representations. Methods Mol Biol 2018;1807:211-24. [Crossref] [PubMed]
- de Hoon MJ, Imoto S, Nolan J, et al. Open source clustering software. Bioinformatics 2004;20:1453-4. [Crossref] [PubMed]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics 2004;20:3246-8. [Crossref] [PubMed]
- Hulsen T, de Vlieg J, Alkema W. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 2008;9:488. [Crossref] [PubMed]
- Li J, Chen L, Xiong Y, et al. Knockdown of PD-L1 in Human Gastric Cancer Cells Inhibits Tumor Progression and Improves the Cytotoxic Sensitivity to CIK Therapy. Cell Physiol Biochem 2017;41:907-20. [Crossref] [PubMed]
- Wang J, Sanmamed MF, Datar I, et al. Fibrinogen-like Protein 1 Is a Major Immune Inhibitory Ligand of LAG-3. Cell 2019;176:334-347.e12. [Crossref] [PubMed]
- Demchev V, Malana G, Vangala D, et al. Targeted deletion of fibrinogen like protein 1 reveals a novel role in energy substrate utilization. PLoS One 2013;8:e58084 [Crossref] [PubMed]
- Zhang Y, Qiao HX, Zhou YT, et al. Fibrinogen-like-protein 1 promotes the invasion and metastasis of gastric cancer and is associated with poor prognosis. Mol Med Rep 2018;18:1465-72. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


