
Transcriptome sequencing-based personalized analysis of hepatocellular carcinoma patients with portal vein tumor thrombus
Introduction
Approximately 10 to 60% of hepatocellular carcinomas (HCCs) invade the portal vein system and form a portal vein tumor thrombus (PVTT). The prognosis of HCC patients with PVTT is poor (1,2). HCC with PVTT has different clinical features, different prognosis, and markedly different treatment outcomes (3-5). PVTT is widely regarded as the main route of intrahepatic metastasis of HCC (6). Currently, hypoxia (7), non-coding ribonucleic acids (RNAs) (8), and cancer stem cells (CSCs) (9) have been found to contribute to PVTT development. Further, numerous molecules, including messenger RNAs (mRNAs), such as Chemokine (C-C motif) ligand 20 (CCL20) (7) and RNA polymerase II subunit 5 (RPB5)-mediating protein (RMP) (10), and long non-coding RNAs (lncRNAs), such as plasmacytoma variant translocation 1 (PVT1) (11), intercellular adhesion molecule-1 (ICAM-1)-related (ICR) (9), and hypoxia-inducible factor 1 alpha-antisense RNA 2 (HIF1A-AS2) (8), have been identified and proposed for use as diagnostic and prognostic biomarkers for HCC, and also represent promising targets for HCC therapy. Based on these findings, we sought to apply these molecular biomarkers to individual HCC patients, and to determine the extent to which they may aid the personalized precision therapy of HCC patients with PVTT.
In this study, we performed RNA sequencing (RNA-seq) on 3 HCC patients with PVTT and aimed to characterize the HCC patients with PVTT at the gene level.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jgo-21-162).
Methods
Patients and samples
Tumor tissue specimens were acquired from 3 Chinese HCC patients who underwent surgery at Sun Yat-sen University Cancer Centre between 2019 and 2020 (Patient 1, female, 53 years; Patient 2, male, 50 years; Patient 3, male, 57 years). Each patient had three matched samples: (I) primary HCC tumor (PT); (II) adjacent normal (N) liver; and (III) PVTT tissues. This study was approved by the Sun Yat-sen University Cancer Centre Institutional Review Board (No. B2020-265). All participants provided written informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
RNA sequencing analysis
Total RNA samples were treated with the RiboMinus Eukaryote Kit (Qiagen, Valencia, CA) to remove ribosomal RNA (rRNA). The NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Beverly, MA) was used to construct the strand-specific RNA-seq libraries in accordance with the manufacturer’s instructions. Briefly, treated RNA samples were fragmented, and then complementary deoxyribonucleic acid (cDNA) were synthesized using random hexamer primers. To remove the second strand of cDNA, a mixture of 2’-Deoxyuridine 5’-Triphosphate (dUTP) was used for synthetic reactions. The End-It DNA End Repair Kit was used to repair cDNA fragments at the ends. These repaired cDNA was then modified with Klenow to add an A at the 3’ end and then linked to adaptors. The linked cDNA products were purified and the second cDNA strand was removed by uracil DNA glycosylase. After purification, the first strand of cDNA was amplified by polymerase chain reaction for 13–16 cycles. The quality of libraries were assessed by Bioanalyzer 2100 (Agilent, Santa Clara, CA), and sequenced by Novaseq S4 PE150 (Illumina, San Diego, CA) with a length of 150 bp.
We aligned the RNA-seq reads to human reference rRNA database using SortMeRNA. Most of the rRNAs were removed by our experiments. Subsequently, the RNA-seq reads were mapped to the human reference genome (hg38) using STAR (Spliced Transcripts Alignment to a Reference) aligner with default parameters. The human genome sequence was downloaded from GENCODE (Homo sapiens GRCh38/hg38). Next, we reassembled a transcriptome using Cufflinks (version 2.2.1) with GENCODE reference annotations for each dataset of the nine samples, based on the mapped reads.
Differential gene expression analysis
We identified genes that were differentially expressed between the PTs or the PVTTs and adjacent normal tissues, and between the PVTTs and the matched PTs using GFOLD (generalized fold change) algorithm for each individual patient. GFOLD calculated its own statistics for expression level changes based on the posterior distribution of the log fold-changes in expression. The significance cutoff was set at 0.01 (-sc 0.01) and |GFOLD value| >1.
TME analysis
CIBERSORT was used to deconvolute the immune microenvironment in each sample (12). Absolute immune cell abundance was calculated from the combat corrected fragments per kilobase per million (FPKM) matrix containing 22 functionally defined human immune subsets (LM22) in the CIBERSORT website.
Statistical analysis
The posterior distribution of the log fold-changes in expression was used to calculate the statistics of GFOLD.
Data and code availability
All the related software and scripts are available from the corresponding author on reasonable request.
Results
Analysis of aberrantly expressed genes
To investigate aberrantly expressed genes in PTs and PVTTs, GFOLD was used to analyze the DEGs between PVTT, PT, and normal tissues from individual patients. Genes with P<0.01 and |GFOLD value| >1 were identified as having significantly different expression. As Figure 1A shows, there were 3,543, 32,472, and 12,901 DEGs between the PT and normal tissues, 5,768, 34,206, and 15,650 DEGs between the PVTT and normal tissues, and 5,015, 23,293, and 22,662 DEGs between the PVTT and PT tissues in Patient 1 (P1), Patient 2 (P2), and Patient 3 (P3), respectively. Among the DEGs, mRNAs accounted for >80% and lncRNAs accounted for 9–14%. The proportion of DEGs between PVTT and PT tissues in P3 (44%) was higher than that of P1 (35%) and P2 (26%). As the Venn diagram shows (see Figure 1B), PVTT and PT tissues shared 455, 18,125, and 2,368 DEGs in P1, P2, and P3, respectively. Among these shared DEGs, 10.5% (48 in 455), 26.9% (4,869 in 18,125), and 59.9% (1,418 in 2,368) of DEGs were differently expressed between PVTTs and PTs. These results indicate that the difference between PVTT and PT in P3 was greater than that in P1 and P2.
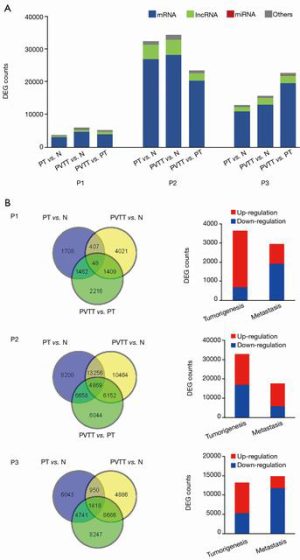
Tumorigenesis-associated and metastasis-associated genes
The number of DEGs between PT and normal tissues were 3,543, 32,472, and 12,901 in P1, P2, and P3, respectively. These DEGs were considered potential tumorigenesis-associated genes. The comparison of PVTTs and PTs detected 5,015, 23,293, and 22,662 DEGs in P1, P2, and P3, respectively, among which 2,919, 17,679, and 14,825 DEGs were differently expressed compared to normal tissues. Thus, these DEGs were considered metastasis-associated genes. As Figure 1B shows, most tumorigenesis-associated genes were upregulated in P1 (81.0%) and P3 (59.6%), and most metastasis-associated genes were downregulated in P1 (66.2%) and P3 (79.2%). Conversely, more tumorigenesis-associated genes were downregulated (52.4%), and more metastasis-associated genes were upregulated (66.3%) in P2.
TME analysis
Based on the cell type–specific gene expression profiles, the proportions of 22 types of immune cells in each sample were calculated using the CIBERSORT algorithm (see Figure 2A). The proportions of M2 macrophages in all 3 patients were >35% in normal tissues, and were 39.7% and 30.7% in PT and PVTT tissues in P1, and 41.6% and 49.7% in PT and PVTT tissues in P2; however, it was greatly decreased in PT tissues (15%) and PVTT tissues (2%) in P3. The proportions of all macrophages (M0, M1, and M2) in N, PT, and PVTT tissues were 42.0%, 45.6%, and 33.0% in P1, 44.2%, 71.2%, and 63.1% in P2, and 37.7%, 17.4%, and 21.5% in P3, respectively (see Figure 2B). The proportions of CD4 T cells in N, PT and PVTT tissues were 14.5%, 17.2%, and 12.1% in P1, 9.3%, 0.9%, and 10.6% in P2, and 21.0%, 24.3%, and 23.3% in P3, respectively (see Figure 2B). The results showed that P2 had the highest proportion of macrophages and the lowest proportion of CD4 T cells, Conversely, P3 had the lowest proportion of macrophages and the highest proportion of CD4 T cells.
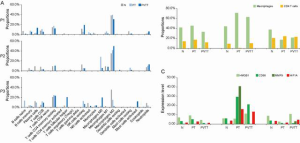
According to previous studies, macrophages are recruited into HCC tissues by upregulating high mobility group box protein 1 (HMGB1) (13), and Cluster of Differentiation 68 (CD68) is frequently used as an indicator for tumor-associated macrophages (TAMs) (14). Moreover, M2 macrophages are reported to promote HCC cell invasion and migration via microRNA (miR)-149-5p/matrix metallopeptidase 9 (MMP9) signaling (15), and the polarization of M2 macrophage can be mediated by HIF1A (16). As Figure 2C shows, the expression level of HMGB1 was lowest in the PT tissue of P3, which is consistent with its having the lowest proportion of macrophages. Similarly, the expression levels of CD68 and MMP9 in the PT tissue of P2, which had the highest proportion of macrophages, were the highest. Further, we found that the expression level of HIF1A was consistent with the proportion of macrophages (see Figure 2B,C), which suggests an interaction between hypoxic microenvironment and macrophage enrichment.
CSCs
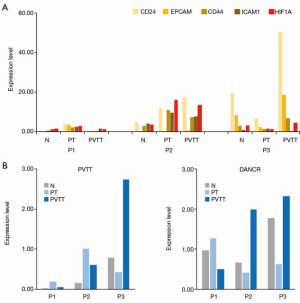
Liver CSCs (LCSCs) play critical roles in cancer initiation, metastasis, recurrence, and therapeutic resistance (17). LCSC surface markers are widely used to identify LCSCs, including epithelial cell adhesion molecules (EPCAM), cluster of differentiation (CD) antigens (CD24, CD133, CD90, CD13, CD44), oval cell marker (OV-6), aldehyde dehydrogenase 1 (ALDH), Keratin 19 (K19), and ICAM-1. The expression level of 4 LCSC surface markers (i.e., CD24, EPCAM, CD44, and ICAM-1) were detected in this study (see Figure 3A). Compared with the PT tissues, PVTT tissues expressed upregulated LCSC surface markers in P2 (CD24) and P3 (CD24, EPCAM and CD44). However, compared with normal tissue, the expression levels of the LCSC surface markers in the PT tissues of P3 were decreased, but were elevated in P1 and P2. Similarly, the expression levels of LCSC surface markers and HIF1A were also consistent, which suggests that the hypoxic nature of the tumor microenvironment (TME) contributes to the increased proportion of LCSC in HCC cells (18).
LncRNAs
Several lncRNAs have been reported to be involved in the tumorigenesis and metastasis of HCC (19), including Highly Up-regulated in Liver Cancer (HULC) (20), delta-like 1 homolog (Drosophila) (DLK1) (21), PVT1 (11), ICR (9), High Expression In HCC (HEIH) (22), microvascular invasion in HCC (MVIH) (23), HIF1A-AS2 (8), and RP11-166D19.1 (8). PVT1 and differentiation antagonizing non-protein coding RNA (DANCR) are stemness-associated lncRNAs. Compared with PT tissues, PVTT tissues expressed upregulated PVT1 in P3 but downregulated PVT1 in P1 and P2, and PVTT tissues expressed upregulated DANCR in P2 and P3, but downregulated DANCR in P1 (see Figure 3B). Based on the results of LCSC surface markers and stemness-associated lncRNAs, we found that P2 and P3 showed stem cell-like properties, but P1 did not.
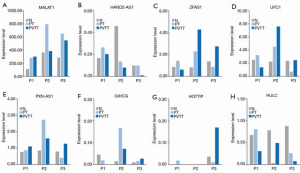
The overexpression of Metastasis-Associated Lung Adenocarcinoma Transcript (MALAT1) promotes proliferation and metastasis in HCC. In all 3 patients, the expression of MALAT1 was upregulated in PT and PVTT tissues compared to that of normal tissues (see Figure 4A). Thus, MALAT1 can be used as universal prognostic markers in HCC. In addition, downregulated lncRNA Heart and Neural Crest Derivatives Expressed 2 antisense RNA (HAND2-AS1) in HCC cells was related to cell migration, and we observed the downregulation of HAND2-AS1 in PVTT compared with PT in all 3 patients (see Figure 4B). In P2, the expression levels of proliferation-associated lncRNAs (i.e., zinc finger nuclear transcription factor, X-box binding 1-type containing 1 antisense RNA 1 (ZFAS1), ubiquitin-fold modifier conjugating enzyme 1 (UFC1), paxillin-antisense RNA 1 (PXN-AS1), and gradually increased during hepatocarcinogenesis (GIHCG)) in the PT and PVTT tissues were much higher than those in normal tissues (see Figure 4C,D,E,F). Moreover, the glutaminolysis-associated lncRNA HOXA transcript at the distal tip (HOTTIP) was overexpressed in the PVTT tissues in P3 (see Figure 4G). The proliferation and lipogenesis-associated lncRNA HULC was not upregulated in PT or PVTT tissues in any patient; thus, HULC does not appear to play an important role in HCC development (see Figure 4H).
Effect of aberrantly expressed genes on survival of HCC patients with PVTT
The progression-free survival (PFS) was 2.8, 3.2, and 3.5 months, and the overall survival (OS) was 7.1, 5.9, and 7.0 months for P1, P2 and P3, respectively. As Figure 5A shows, the PFS was comparable in all 3 patients, but P2 had the shortest OS, and P3 had the longest OS. According to the results (see above), the expression level of HIF1A was greatly elevated in PT tissues (which was 4.5 times of the normal tissue) and PVTT tissues (which was 3.8 times that of normal tissue) in P2 (see Figure 5B). Accordingly, the expression level of CCL20 was elevated in PT tissues (which was 1.6 times that of normal tissue) and PVTT tissues (which was 11.2 times that of normal tissue) (see Figure 5C), and the expression level of indoleamine 2, 3-dioxygenase (IDO) was also elevated in PVTT tissues (see Figure 5D). Additionally, compared with normal tissue (44.2%), macrophages were recruited into PT tissues (71.2%) and PVTT tissues (63.1%) in P2 (see Figure 2B), suggesting that hypoxia induces an immunosuppressive TME and promotes tumorigenesis and metastasis.
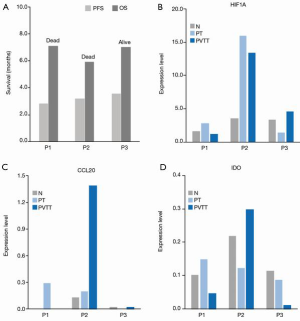
In P3, 3 LCSC surface markers were greatly overexpressed in PVTT tissues, including CD24 (which was 7.5 times that of the PT tissues), EPCAM (which was 8.1 times that of the PT tissues) and CD44 (which was 4.8 times that of the PT tissues) (see Figure 3A). Moreover, 2 stemness-associated lncRNAs (i.e., PVT1 and DANCR) were upregulated 6.5 and 3.7 times, respectively, in PVTT tissues compared to that of PT tissues (see Figure 3B). Thus, CSCs appeared to play critical roles in metastasis in P3.
In P1, compared with normal tissue, the expression of HIF1A was upregulated in PT tissues but downregulated in PVTT tissues (see Figure 5B). The same trend was observed in the expression of LCSC surface markers (see Figure 3A) and stemness-associated lncRNAs (see Figure 3B). In addition, the expression level of other lncRNAs changed slightly. The results indicated that hypoxia and CSCs may contribute to the tumorigenesis but not to the metastasis.
Discussion
The management of HCC patients with PVTT is challenging. To determine the best treatment strategy for patients with PVTT, HCC patients with PVTT need to be classified to guide treatment and to compare outcomes between different groups. At present, different classification systems have been developed at different centers. The most conventional classification for PVTT was proposed by the Liver Cancer Study Group of Japan (LCSGJ) (24): Vp0 represents no PVTT; Vp represents the presence of PVTT distal to, but not in, the secondary portal vein branches; Vp2 represents the presence of PVTT in the secondary portal vein branches; Vp3 represents the presence of PVTT in the primary portal vein branch; and Vp4 represents the presence of PVTT in the main portal vein or contralateral portal vein branch or both.
Shi et al. (4) classified HCC with PVTT into four types: Type 1—PVTT involving segmental branches or above; Type 2—PVTT involving the right or left portal vein; Type 3—PVTT involving the main portal vein; and Type 4—PVTT involving the superior mesenteric vein. Xu et al. (25) divided PVTT patients into two groups: Group A—PVTT involving the main portal vein trunk or both the left and right portal veins, and Group B—PVTT involving only the left or right portal vein. However, a molecular classification of HCC patients with PVTT has not been developed yet.
Many studies have focused on the mechanism of PVTT in HCC at the gene level, and identified many biomarkers for tumorigenesis and metastasis (7-9,17,19). To promote these findings for clinical application, we performed RNA-seq on 3 HCC patients with PVTT to determine whether these biomarkers can help to stratify HCC patients with PVTT, and aid in the personalizing of precision therapy. We found that tumor development and metastasis in the 3 patients were affected by different mechanisms. Hypoxia and CSCs may have contributed to tumorigenesis but not to metastasis in P1. Hypoxia microenvironment played an important role in tumorigenesis and metastasis in P2, and CSCs may have contributed to the metastasis. CSCs played critical roles in metastasis but not in tumorigenesis in P3. We also found that lncRNA MALAT1 was greatly overexpressed in PT and PVTT tissues in all 3 patients, and HAND2-AS1 was downregulated in PVTT compared with PT tissues in all 3 patients, which suggests that MALAT1 and HAND2-AS1 may be robust biomarkers for metastasis in HCC with PVTT. In addition, HULC may not play any role in tumorigenesis or metastasis in HCC patients with PVTT. Other lncRNAs, including ZFAS1, UFC1, PXN-AS1, GIHCG, and HOTTIP, produced inconsistent changes in the expression levels in the 3 patients.
We also found that the proportion of macrophages in the microenvironment was closely related to the survival of HCC patients with PVTT. The more the macrophages in the TME, the shorter the OS. As reported previously, while macrophages are essential for the normal activity of the immune system, their aberrant regulations are related to HCC (26). Macrophages largely infiltrate HCC microenvironment and play roles in the development and progression of HCC, including immunosuppressive function, enhancement of tumor invasion and metastasis, angiogenesis, epithelial-mesenchymal transition, and maintenance of stemness. Thus, there is considerable interest in TAMs targeted immunotherapy for HCC. These strategies include inhibition of monocyte recruitment, elimination of TAMs that are already present in tumor tissues, functional re-education of TAMs polarization, and neutralization of tumor-promoting products of TAMs. Preliminary data on TAMs-targeted drug interventions suggest that these insights can be successfully translated into new treatment options for HCC patients.
Conclusions
As HCC patients with PVTT are heterogeneous at the gene level, driven by different mechanisms, and have different prognoses, individualized management strategies are required. TAMs-targeted immunotherapy represents a promising therapy for HCC patients with PVTT. Further, MALAT1 and HAND2-AS1 represent promising targets for HCC therapy.
Acknowledgments
We would like to thank Shanghai Tongshu Biotechnology Co. Ltd. for providing us with technical support, and all of the participants for their kind cooperation in this study.
Funding: This study was funded by the National Natural Science Foundation of China (Grant No. 81571780).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-21-162
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-21-162
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-21-162). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Sun Yat-sen University Cancer Centre Institutional Review Board (No. B2020-265). All participants provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Quirk M, Kim YH, Saab S, et al. Management of hepatocellular carcinoma with portal vein thrombosis. World J Gastroenterol 2015;21:3462-71. [Crossref] [PubMed]
- Zhong JH, Peng NF, You XM, et al. Tumor stage and primary treatment of hepatocellular carcinoma at a large tertiary hospital in China: A real-world study. Oncotarget 2017;8:18296-302. [Crossref] [PubMed]
- Shuqun C, Mengchao W, Han C, et al. Tumor thrombus types influence the prognosis of hepatocellular carcinoma with the tumor thrombi in the portal vein. Hepatogastroenterology 2007;54:499-502. [PubMed]
- Shi J, Lai EC, Li N, et al. A new classification for hepatocellular carcinoma with portal vein tumor thrombus. J Hepatobiliary Pancreat Sci 2011;18:74-80. [Crossref] [PubMed]
- Katagiri S, Yamamoto M. Multidisciplinary treatments for hepatocellular carcinoma with major portal vein tumor thrombus. Surg Today 2014;44:219-26. [Crossref] [PubMed]
- Mitsunobu M, Toyosaka A, Oriyama T, et al. Intrahepatic metastases in hepatocellular carcinoma: the role of the portal vein as an efferent vessel. Clin Exp Metastasis 1996;14:520-9. [Crossref] [PubMed]
- Ye LY, Chen W, Bai XL, et al. Hypoxia-Induced Epithelial-to-Mesenchymal Transition in Hepatocellular Carcinoma Induces an Immunosuppressive Tumor Microenvironment to Promote Metastasis. Cancer Res 2016;76:818-30. [Crossref] [PubMed]
- Yang Y, Chen L, Gu J, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun 2017;8:14421. [Crossref] [PubMed]
- Guo W, Liu S, Cheng Y, et al. ICAM-1-Related Noncoding RNA in Cancer Stem Cells Maintains ICAM-1 Expression in Hepatocellular Carcinoma. Clin Cancer Res 2016;22:2041-50. [Crossref] [PubMed]
- Zhang J, Pan YF, Ding ZW, et al. RMP promotes venous metastases of hepatocellular carcinoma through promoting IL-6 transcription. Oncogene 2015;34:1575-83. [Crossref] [PubMed]
- Wang F, Yuan JH, Wang SB, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology 2014;60:1278-90. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Jiang J, Wang GZ, Wang Y, et al. Hypoxia-induced HMGB1 expression of HCC promotes tumor invasiveness and metastasis via regulating macrophage-derived IL-6. Exp Cell Res 2018;367:81-8. [Crossref] [PubMed]
- Chistiakov DA, Killingsworth MC, Myasoedova VA, et al. CD68/macrosialin: not just a histochemical marker. Lab Invest 2017;97:4-13. [Crossref] [PubMed]
- Liu G, Yin L, Ouyang X, et al. M2 Macrophages Promote HCC Cells Invasion and Migration via miR-149-5p/MMP9 Signaling. J Cancer 2020;11:1277-87. [Crossref] [PubMed]
- Ambade A, Satishchandran A, Saha B, et al. Hepatocellular carcinoma is accelerated by NASH involving M2 macrophage polarization mediated by hif-1αinduced IL-10. Oncoimmunology 2016;5:e1221557 [Crossref] [PubMed]
- Wang N, Wang S, Li MY, et al. Cancer stem cells in hepatocellular carcinoma: an overview and promising therapeutic strategies. Ther Adv Med Oncol 2018;10:1758835918816287 [Crossref] [PubMed]
- Muramatsu S, Tanaka S, Mogushi K, et al. Visualization of stem cell features in human hepatocellular carcinoma reveals in vivo significance of tumor-host interaction and clinical course. Hepatology 2013;58:218-28. [Crossref] [PubMed]
- Huang Z, Zhou JK, Peng Y, et al. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer 2020;19:77. [Crossref] [PubMed]
- Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007;132:330-42. [Crossref] [PubMed]
- Huang J, Zhang X, Zhang M, et al. Up-regulation of DLK1 as an imprinted gene could contribute to human hepatocellular carcinoma. Carcinogenesis 2007;28:1094-103. [Crossref] [PubMed]
- Yang F, Zhang L, Huo XS, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011;54:1679-89. [Crossref] [PubMed]
- Yuan SX, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology 2012;56:2231-41. [Crossref] [PubMed]
- Kudo M, Kitano M, Sakurai T, et al. General Rules for the Clinical and Pathological Study of Primary Liver Cancer, Nationwide Follow-Up Survey and Clinical Practice Guidelines: The Outstanding Achievements of the Liver Cancer Study Group of Japan. Dig Dis 2015;33:765-70. [Crossref] [PubMed]
- Xu JF, Liu XY, Wang S, et al. Surgical treatment for hepatocellular carcinoma with portal vein tumor thrombus: a novel classification. World J Surg Oncol 2015;13:86. [Crossref] [PubMed]
- Tian Z, Hou X, Liu W, et al. Macrophages and hepatocellular carcinoma. Cell Biosci 2019;9:79. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


