
KRAS gene status in gastric signet-ring cell carcinoma patients and acts as biomarker of MEK inhibitor
Introduction
Gastric cancer (GC) is the fifth most common cancer in the world and currently accounts for 8.2% of all new cancer cases worldwide (1-3). Signet-ring cell carcinoma (SRCC) is defined as GC with at least 50% of signet-ring cell in the pathologic specimen according to World Health Organization (WHO) classification and is found in 8% to 30% of GC (4). Distinct epidemiology and oncogenesis of SRCC have been observed in previous studies. For example, SRCC occurs more often in younger women groups and SRCC is associated with serosal invasion, lymph node metastasis and worse prognosis (4,5). Although The Cancer Genome Atlas (TCGA) has systematical analysis about four subtypes of GC through next generation sequencing. Few studies have been focused on SRCC molecular subtypes, and a significant proportion of clinical regimens are limited to chemotherapy.
KRAS is renowned as one of the most common driven genes in cancer. Research on RAS-driven cancers almost focused on RAS-coding mutations. Ninety-nine percent of cancer-associated KRAS genes mutations are characterized by single base missense mutations, which are found at residues G12, G13 or Q61 (6). In colorectal, pancreatic, and non-small cell lung cancers (NSCLCs), KRAS mutations have been found to have a prognostic impact (6-9). Other measures for KRAS alterations, although less studied than mutations, also plays an important role. Recently, studies suggest that amplification and overexpression of KRAS was associated with enhanced rates of metastasis or poor survival in ovarian and endometrial cancers (10,11). However, Ras proteins have been dismissed as “undruggable” for many years. Till now, only one therapeutic attack AMG510 had been yielded. AMG 510, which targets a KRAS mutation known as G12C was reported safety, tolerability and preliminary anti-tumour activity in a phase I trial (12).
More attention has been paid to downstream pathways of RAS, especially the RAS-RAF-MAPK pathway and the PI3k-Akt-mTor kinase pathway. The goal of inhibiting these pathways is to provide clinical benefit to patients with RAS driven cancers (13).
In this study, we systematically investigated KRAS gene alterations in TCGA database. We also investigated the association between KRAS gene status and clinical outcomes. Furthermore, sensitivity and response of 4 SRCC cell lines to MEK and mTOR inhibitors were also determined.
We present the study in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-617).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the ethics committee of Drum Tower Hospital (No. 2016-196-01), and informed consent was obtained from all subjects.
Bioinformatics analysis
We used publicly available data from TCGA in this study. Clinical information, mRNA expression and gene mutation data from TCGA-STAD cohort contained 388 tumor samples were downloaded from the NCI’s Genomic Data Commons (GDC) portal (https://portal.gdc.cancer.gov). Raw RNA-seq count matrix and clinical information were obtained using RTCGAToolbox (2.8.0) package in R (3.4.4). The count matrix was normalized and processed by log2 transformation using Deseq2 (1.18.1) package. Subsequently, statistical analyses were performed to evaluate the KRAS expression in STAD tissues and the correlation between KRAS expression and clinical outcomes. Additionally, to further analyze the overall survival of STAD patients, a Kaplan-Meier plotter was constructed according to the median KRAS expression value or KRAS alteration status.
Patients and samples
This study included 234 SRCC patients and 57 intestinal gastric adenocarcinoma patients who underwent gastrectomy at the General Surgery Department of Drum Tower Hospital between 2010 and 2016. All patients were pathologically confirmed by pathologist and the clinical data were collected from medical charts and pathology reports.
Immunohistochemistry
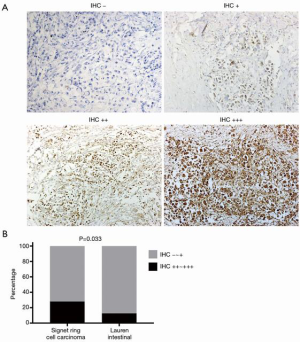
Seventy-five SRCC patients and 57 intestinal gastric adenocarcinoma patients in our internal cohort which had sufficient tumor tissues were fixed in formalin and embedded in paraffin. Sections of a thickness of 4 µm were obtained from the tissue paraffin blocks and used for IHC. Paraffin sections were deparaffinized in xylene and rehydrated in a graded alcohol series and distilled water. The slides were heated for antigen retrieval in 1 mmol/L EDTA (0.05% Tween 20, pH 8.0). Endogenous peroxidase was blocked by incubation with 3% H2O2 for 10 min. Then the sections were blocked with 10% goat serum at room temperature for 1 h. After washing with PBS, anti-KRAS (ab180772, Abcam, Cambridge, UK) antibody (1:100 dilution) was applied to the sections, and the sections were incubated at 4 °C overnight. The combination of primary antibody was detected by peroxidase staining with an avidin-biotin complex system. KRAS expression was evaluated semi-quantitatively according to the degree and proportion of membrane staining at the same time: 0 manifested no staining is observed or membrane staining is observed in <10% of tumor cells; 1+ mean faint or partly membrane staining is found in >10% of tumor cells; 2+ represented weak to moderate complete membrane staining is detected in >10% of tumor cells; 3+ represented strong, complete membrane staining is observed in >10% of tumor cells
KRAS sequence
KRAS exon 2 and exon 3 mutations were identified by PCR on 50 ng DNA samples. The primers used were summarized in Table 1. The Sanger sequencing process was performed by STAB Vida (Caparica, Portugal). PCR products were then directly sequenced using the Applied Biosystems (ABI) PRISM 3730 XL. PCR amplification was repeated to confirm the results.
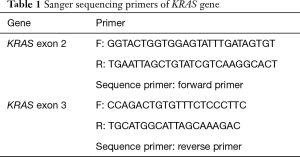
Full table
FISH for KRAS gene amplification
Four-micron-thick FFPE tissue sections were used for FISH testing. Hematoxylin and eosin (H&E)-stained score was determined by two independent observers who were blinded to the clinicopathological information of each sample. KRAS gene copy number was then investigated according to protocol from Abbot molecular (Abbot molecular, Wiesbaden, Germany). Briefly, slides were incubated at 56 °C overnight, deparaffinized, and hydrated. After incubation in 2× saline sodium citrate buffer (pH =7.0) at 75 °C for 30 min slides were digested with 20 µg/mL proteinase K in pre-warmed 50 mM Tris for 10–20 min at 37 °C. Then rinse the slides 5 times in distilled water and then immerse slides in ice-cold 2× saline sodium citrate buffer for 20 s. After that, slides were dehydrated using ethanol in a series of increasing concentrations. Hybridization was performed in a humidified chamber at 37 °C for 14–18 h with a denatured DNA probe onto the selected area based on the presence of tumor foci on each slide. Finally, the slides were air dried in the dark and counterstained with 4,6-diamidino-2-phenylindole (DAPI). FISH signals were assessed with a Zeiss fluorescence microscope (Gottingen, Germany) by two independent investigators. The numbers of KRAS probe signals and CHR12 CEP probe (CEP12) signals were counted for each nucleus, and an overall mean KRAS:CEP12 ratio was calculated for each patient. Each nucleus was classified according to the number of copies of KRAS and CEP12. Gene amplification was defined as a ratio of KRAS to CEP12 of ≥2, polysomy was defined as CEP12 ≥3, disomy (normal) was defined by CEP12 and KRAS signal =2, as previously described (14). Results were interpreted by a pathologist and a cytogeneticist and were reported using ASCO/CAP 2013 criteria.
Cell lines and culture conditions
KATO-III cell lines were obtained from the Chinese Collection of Research Bioresources. Other cell lines (SNU601, SNU668, and NUGC-4) were provided by the National Cancer Center Research Institute (Tokyo, Japan) in 2011. Cells were cultured in RPMI-1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (Gibco, NY, USA) at 37 °C in a humidified atmosphere of 5% CO2.
Drug treatment and cell viability assay
AZD6244 and AZD2014 were purchased from Selleck (Houston TX, USA). Cells were seeded in 96-well plates (3,000 cells per well) with antibiotic-free RPMI 1640 (Invitrogen) plus 10% fetal bovine serum at 37 °C with 5% CO2 for 24 h. The cells were then treated with AZD6244 and AZD2014 for another 72 h to determine the 50% inhibition concentrations (IC50). The growth-inhibitory effects of AZD6244 and AZD2014 were tested by 3,4,5-dimethyl-2H-tetrazolium bromide assay (MTT; Sigma-Aldrich). Optical density was spectrophotometrically measured at 570 nM. Each experiment was carried out in triplicate and data are presented as geometric means.
Western blotting
Western blotting analysis was performed in four gastric cell lines as described previously (15). Cells were lysed in buffer containing 50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.1% SDS, 1% Nonidet P-40, and 0.5% sodium-deoxycholate. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto to PVDF membranes. After blocked with 10% defatted milk, the membranes were incubated overnight at 4 °C with anti-c-K-Ras (1:1,000, Millipore) antibodies. After further washes, the membranes were incubated with the goat anti-rabbit/mouse peroxidase-conjugated secondary antibodies (Abcam), and the blots were developed using ECL (Millipore).
Statistical analysis
Data was analyzed using independent two-tailed t-test. Categorical data was analyzed using the two-side chi-square test. The survival distributions were obtained by the Kaplan-Meier survival analysis and compared using the log-rank test, considering death for cancer as the end point. All statistical calculations were performed with the Statistical Package for the Social Sciences for Windows version 22 (SPSS Inc., Chicago, IL, USA). Two-sided P values <0.05 were considered to indicate statistical significance.
Results
TCGA analysis
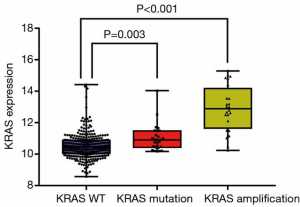
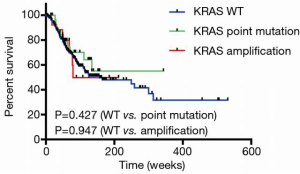
Genomic mutation, copy number variants and RNA expression data were extracted from TCGA. Patients with complete pathologic and genomic data were included for further analysis. Their characteristics were summarized in Table 2. There are 388 patients including 256 men and 132 women and the median age was 66. There are 8 specific histological types in the TCGA STAD dataset. Among the 388 patients, only 11 patients were pathologically diagnosed as SRCC. For tumor stage, 105 patients and 276 patients were diagnosed with T1–2 and T3–4 respectively. Two hundred and sixty patients had lymph nodes metastasis (N1–3) and 26 patients got distant metastasis (M1). Thirty patients (7.7%) harbored point mutations, largely [26/30] showed exon 12 or exon 13 mutations, which was consistent with previous studies. Thirty-three patients (8.5%) had more than one copy number amplification. Two of the patients had both KRAS point mutation and amplification. The majority of the patients [21/33] gained more than 12 copies amplification. We further investigated the relationship between KRAS alteration and its expression. Interestingly, patients harboring KRAS point mutations and copy number variation (CNV) showed higher mRNA level compared to non-mutant cases (P=0.003 and P<0.001) (Figure 1). Kaplan-Meier analyses of the entire patient cohort showed that KRAS alteration status had no significant prognostic value for patients' survival (Figure 2).

Full table
Internal cohort analysis
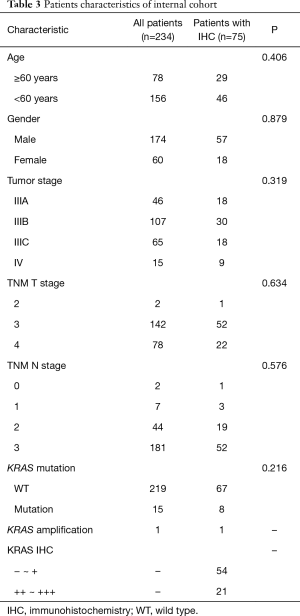
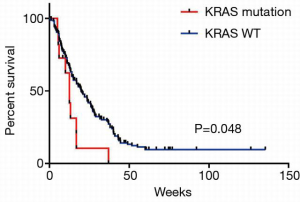
We further analyzed our internal cohort. Among them, 234 cases were histologically confirmed SRCC. The median age was 58.26 years old (range, 33–88 years old). Patients’ characteristics were summarized in Table 3. Fifteen patients were detected with KRAS mutations. There were 8 G12V, 3 G12D, 1 G12S, 1 G12C, 1 G13D, and 1 G151A. Survival analysis showed that these patients had a significantly lower OS (Figure 3, P=0.048). We further did analysis on the 75 samples which had sufficient tumor tissues. In these 75 cases, 8 showed KRAS mutations with 7 G12V and 1 G151A. Two cases showed KRAS amplification. KRAS protein expression was assessed by IHC and the results showed that 54 patients were detected with negative (−) or weak positive (+) expression, whereas 21 patients were KRAS median (++) or strong positive (+++). To investigate KRAS expression in GC, we next explored KRAS expression in another internal cohort which was consisted of 57 intestinal gastric adenocarcinoma patients. In intestinal cohort, 50 patients were detected with negative (−) or weak positive (+) expression, 7 patients were KRAS median (++) or strong positive (+++). Obviously, the majority of SRCC patients were median or strong positive KRAS expression, which is higher than our intestinal cohort (28% vs. 12.6%, P=0.033, Figure 4). Survival analysis showed that the median OS was 12.5 months for patients with KRAS mutation, and 19.5 months for patients without KRAS mutation (P=0.005, Figure 5A). However, different KRAS expression levels had no effect on OS (P=0.095, Figure 5B).
Full table

Cell line investigations
To investigate the drug response of SRCC cell lines according to KRAS gene status, the cytotoxicity of MEK and mTOR inhibitors in four gastric SRCC cancer cell lines (SNU601, SNU668, KATO-III and NUGC-4) was assessed by MTT assay. KRAS alteration in cell lines according to sanger sequence was summarized in Table 4. Three cell lines, which were KATO-III, SNU-601 and NUGC-4, showed detectable KRAS expression (Figure 6). However, none of the cell lines showed KRAS amplification. All four cell lines were treated with MEK inhibitor AZD6244 and mTOR inhibitor AZD2014 for 72 h. SNU601 and SNU668, which harbored KRAS mutation, were more hypersensitive to AZD6244 and AZD2014 than other cell lines. They showed significantly lower IC50 than cells without KRAS mutations (Table 4).

Full table
Discussion
GC can be classified histologically into various types and SRCC is defined as cells with abundant intracytoplasmic mucin (16). SRCC is associated with more aggressive cancer and the disease is usually diagnosed at a distant stage (17). Although almost all SRCC belonged to diffused type, distinct epidemiology and oncogenesis of SRCC have been observed (5).
Gene alteration of KRAS has considered to be an important biological biomarker in several cancer types. Oncogenic Ras proteins promote multiple cancer related event, including excessive proliferation, loss of contact inhibition, increased motility, and so on (18). We firstly systematically investigated the KRAS gene alteration including point mutations and CNV in 389 patients in TCGA database. However, only 11 patients were pathologically diagnosed as SRCC. None of these 11 SRCC patients harbored KRAS point mutations or CNV, which made it unavailable for further subtype analysis. Regretfully, no survival difference was observed between different KRAS gene status in TCGA database. In consideration of the lower prevalence of SRCC, our cohort included 234 SRCC patients was selected from thousands of GC samples. Previous studies have shown that SRCC tend to occur more often in younger women groups. However, 60/234 (25.6%) patients were women in our cohort. Our cohort was selected from GC surgery samples. Due to the advanced clinical stage, the majority of patients might not receive surgical treatment, resulting in a gender bias. In our internal SRCC cohort, 6.4% patients harbored KRAS point mutation in exon 2 and exon 3 which was consistent with previous report identifying 6.5% KRAS mutation rate in GC (19).
KRAS is the most frequently mutated oncogene compared with other RAS subtypes (NRAS, HRAS). KRAS mutations was previously mainly studied in endometrial cancer, however, the prognostic importance of KRAS mutation was inconsistent (11,20). Studies showed that KRAS mutations were associated with inferior survival in colon cancer patients (21,22) and in metastasis pancreatic cancer (23). Mutations in codons 12 and 13 are correlated with poor OS for colorectal cancer patients (24,25) and are predictors of resistance or sensitivity to EGFR-targeted therapy (26-28) by activated both the PI3K/AKT and MAPK signaling (29). Interestingly, we found that KRAS mutation were associated with unfavorable disease survival. We also found a higher KRAS expression level in SRCC cohort than our intestinal cohort (80% vs. 38.6%, P<0.001). To the best of our knowledge, this is the first study to demonstrate that KRAS expression was more common in SRCC and that KRAS mutation independently predicts worse OS in SRCC patients. Previous studies reported that KRAS mutations with MSS status presented a poor prognosis and a worse outcome (30). KRAS G12V mutation carriers have much shorter OS than other mutation carriers and wild-type groups (31). However, our study is a retrospective analysis, large prospective studies might be needed to validate our findings.
KRAS was long thought to be undruggable because the protein lacked traditional small molecule binding pockets. In 2019, AMG 510, which inhibits the oncogene KRAS, has achieved something which has eluded many other drug makers. The study of AMG 510 in 35 patients found no dose-limiting toxicities at the tested dose levels, and revealed a 50% partial response rate among a subgroup of 10 patients with KRAS-positive NSCLC (12). Despite more than three decades of effort, no effective Ras inhibitors have been approved. Thus, the question was raised about which pathway should be targeted. Though the MAPK pathway is preferentially activated by RAS, it has been also known to interact with p110α, the catalytic subunit of PI3K and PI3K signaling is firmly established under RAS (32). The direct downstream of the RAS signaling are the MAPK and PI3K signaling pathways, and MEK and mTOR inhibitors block these two pathways (33). More efforts have been put into MAPK, MEK and mTOR inhibitors. A pre-clinical study demonstrated that MAPK inhibitor showed superior efficacy in GC cell lines with KRAS mutation. In previous study, MEK inhibition showed significant treatment effects on KRAS-dependent CRC (34). Furthermore, MEK inhibitor could reduce peritoneal dissemination of KRAS addicted ovarian cancer. In addition, combined inhibition of the PI3K/mTOR/MEK pathway could induces apoptosis in pancreatic cancer cells (35). In our study, KRAS mutation was associated with increased sensitivity to MEK and mTOR inhibitors in gastric SRCC cell lines which was in line with the previous studies. Further studies also focused on tyrosine phosphatase SHP2, which was critical for mutant KRAS-driven cancers, and necessary for resistance mechanisms upon blockade of MEK (36). These results, together with our study, suggest that MEK and mTOR inhibitor might be a promising therapeutic options of signet ring cell GC with KRAS mutation.
Nowadays, new treatment methods for KRAS mutation are finally coming into view (37). Due to mutant KRAS proteins themselves are not strong antigens, efforts are now focus on increasing the capacity of the immune system to recognize KRAS mutants as neoantigens. Since this perspective has been aroused for many years, progress had been made in generating T-cell responses against Ras-derived epitopes (18). Immune checkpoint molecules play an important role in tumor immune escape, and the recent development of immune checkpoint inhibitors showed sufficient efficacy of anti-PD-1 antibody in specific subset of NSCLC patients. A recent pre-clinical study demonstrated that PD-L1 expression was positively associated with KRAS mutation in both KRAS mutated NSCLS cell lines and tissues. Further investigation revealed that PD-L1 was up-regulated by KRAS mutation through p-ERK signaling (38). The relationship of KRAS and PD-1 was also verified in clinical studies. In a retrospective study, advanced NSCLC patients treated with a single checkpoint inhibitor had at least one oncogenic driver change, patients with KRAS mutation showed more long-term response (12 months), higher partial or complete response rate and lower rapid progression rate (within 2 months) compared with other mutations (such as EGFR, BRAF, MET, HER2, ALK, RET, and ROS1 mutations) (39). However, in colorectal cancer, KRAS mutation reduced T cell infiltration and anti-PD-1 therapy resistance in mouse model (40). These data indicate potential role of immunotherapy in selected KRAS mutation.
In conclusion, our findings show for the first time that KRAS mutation is a promising prognostic marker in SRCC patients. Furthermore, KRAS mutation can be used as a predictive marker in patients treated with MEK and mTOR inhibitors. Given that SRCC was not sensitive to common chemotherapeutic agents, the results our study may facilitate further development of agents targeted agents in SRCC.
Acknowledgments
Funding: This study was supported financially by grants from the Ministry of Science and Technology of the People’s Republic of China (2019ZX09301-150) and grants from Department of Science and Technology of Jiangsu Province (BK2019001).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-617
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-617
Peer Review File: Available at http://dx.doi.org/10.21037/jgo-20-617
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-617). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experimental methods were carried out in accordance with the approved guidelines. This study was conducted with the approval of the Ethics Committee of Nanjing Drum Tower Hospital (No. 2016-196-01). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and informed consent was taken from all the patients
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Liu X, Cai H, Sheng W, et al. Clinicopathological Characteristics and Survival Outcomes of Primary Signet Ring Cell Carcinoma in the Stomach: Retrospective Analysis of Single Center Database. PLoS One 2015;10:e0144420 [Crossref] [PubMed]
- Pernot S, Voron T, Perkins G, et al. Signet-ring cell carcinoma of the stomach: Impact on prognosis and specific therapeutic challenge. World J Gastroenterol 2015;21:11428-38. [Crossref] [PubMed]
- Cox AD, Fesik SW, Kimmelman AC, et al. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014;13:828-51. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. [Crossref] [PubMed]
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008;83:584-94. [Crossref] [PubMed]
- Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011;39:D945-50. [Crossref] [PubMed]
- Ross JS, Ali SM, Wang K, et al. Comprehensive genomic profiling of epithelial ovarian cancer by next generation sequencing-based diagnostic assay reveals new routes to targeted therapies. Gynecol Oncol 2013;130:554-9. [Crossref] [PubMed]
- Birkeland E, Wik E, Mjos S, et al. KRAS gene amplification and overexpression but not mutation associates with aggressive and metastatic endometrial cancer. Br J Cancer 2012;107:1997-2004. [Crossref] [PubMed]
- Fakih M, O'Neil B, Price TJ, et al. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J Clin Oncol 2019;37:abstr 3003.
- McCormick F. KRAS as a Therapeutic Target. Clin Cancer Res 2015;21:1797-801. [Crossref] [PubMed]
- Sasaki H, Hikosaka Y, Kawano O, et al. Evaluation of Kras gene mutation and copy number gain in non-small cell lung cancer. J Thorac Oncol 2011;6:15-20. [Crossref] [PubMed]
- Dulak AM, Schumacher SE, van Lieshout J, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res 2012;72:4383-93. [Crossref] [PubMed]
- Chu PG, Weiss LM. Immunohistochemical characterization of signet-ring cell carcinomas of the stomach, breast, and colon. Am J Clin Pathol 2004;121:884-92. [Crossref] [PubMed]
- Shu Y, Zhang W, Hou Q, et al. Prognostic significance of frequent CLDN18-ARHGAP26/6 fusion in gastric signet-ring cell cancer. Nat Commun 2018;9:2447. [Crossref] [PubMed]
- Yuan TL, Amzallag A, Bagni R, et al. Differential Effector Engagement by Oncogenic KRAS. Cell Rep 2018;22:1889-902. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Semczuk A, Berbec H, Kostuch M, et al. K-ras gene point mutations in human endometrial carcinomas: correlation with clinicopathological features and patients' outcome. J Cancer Res Clin Oncol 1998;124:695-700. [Crossref] [PubMed]
- Yoon HH, Tougeron D, Shi Q, et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance). Clin Cancer Res 2014;20:3033-43. [Crossref] [PubMed]
- Li W, Qiu T, Zhi W, et al. Colorectal carcinomas with KRAS codon 12 mutation are associated with more advanced tumor stages. BMC Cancer 2015;15:340. [Crossref] [PubMed]
- Rachagani S, Senapati S, Chakraborty S, et al. Activated KrasG(1)(2)D is associated with invasion and metastasis of pancreatic cancer cells through inhibition of E-cadherin. Br J Cancer 2011;104:1038-48. [Crossref] [PubMed]
- Downward J. RAS's cloak of invincibility slips at last? Cancer Cell 2014;25:5-6. [Crossref] [PubMed]
- Hayama T, Hashiguchi Y, Okamoto K, et al. G12V and G12C mutations in the gene KRAS are associated with a poorer prognosis in primary colorectal cancer. Int J Colorectal Dis 2019;34:1491-6. [Crossref] [PubMed]
- Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 2005;23:5900-9. [Crossref] [PubMed]
- Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res 2007;13:2890-6. [Crossref] [PubMed]
- Kumar SS, Price TJ, Mohyieldin O, et al. KRAS G13D Mutation and Sensitivity to Cetuximab or Panitumumab in a Colorectal Cancer Cell Line Model. Gastrointest Cancer Res 2014;7:23-6. [PubMed]
- Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228-39. [Crossref] [PubMed]
- Polom K, Das K, Marrelli D, et al. KRAS Mutation in Gastric Cancer and Prognostication Associated with Microsatellite Instability Status. Pathol Oncol Res 2019;25:333-40. [Crossref] [PubMed]
- Fu XH, Chen ZT, Wang WH, et al. KRAS G12V Mutation is an Adverse Prognostic Factor of Chinese Gastric Cancer Patients. J Cancer 2019;10:821-8. [Crossref] [PubMed]
- Gupta S, Ramjaun AR, Haiko P, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell 2007;129:957-68. [Crossref] [PubMed]
- Castellano E, Downward J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes Cancer 2011;2:261-74. [Crossref] [PubMed]
- Pek M, Yatim S, Chen Y, et al. Oncogenic KRAS-associated gene signature defines co-targeting of CDK4/6 and MEK as a viable therapeutic strategy in colorectal cancer. Oncogene 2017;36:4975-86. [Crossref] [PubMed]
- Burmi RS, Maginn EN, Gabra H, et al. Combined inhibition of the PI3K/mTOR/MEK pathway induces Bim/Mcl-1-regulated apoptosis in pancreatic cancer cells. Cancer Biol Ther 2019;20:21-30. [Crossref] [PubMed]
- Ruess DA, Heynen GJ, Ciecielski KJ, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med 2018;24:954-60. [Crossref] [PubMed]
- Chan TA, Wolchok JD, Snyder A. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med 2015;373:1984. [Crossref] [PubMed]
- Chen N, Fang W, Lin Z, et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother 2017;66:1175-87. [Crossref] [PubMed]
- Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol 2019;30:1321-8. [Crossref] [PubMed]
- Liao W, Overman MJ, Boutin AT, et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell 2019;35:559-572.e7. [Crossref] [PubMed]



