
Integrated analysis of competing endogenous RNA in esophageal carcinoma
Introduction
Esophageal carcinoma (ESCA), is the sixth leading cause of tumor-related death of men worldwide (1). Until now, the ESCA has been known to contain two histologic subtypes; esophageal squamous cell carcinoma (ESCC), and esophageal adenocarcinoma (ECA). It is widely known that the occurrence of these two major histologic subtypes differs among different regions. The ESCC is prevalent in Asia, Southern Europe, and Eastern and Southern Africa, whereas ECA mainly occurs in North America and some regions of Europe (2). In China, ESCA is the fourth most deadly tumor and fifth in incidence among all malignant tumors (3,4) due to poor early diagnostic strategy and aggressively invasive nature of the cancer cells (5). Therefore, it is essential to develop therapeutic targets and novel molecular biomarkers for ESCA.
The competing endogenous RNA (CeRNA) hypothesis has been reported by researchers, elucidating a novel regulatory mechanism between non-coding RNA (ncRNA) and messenger RNA (mRNA) (6). Based on the CeRNA hypothesis, more and more studies have found that interactions between CeRNAs through sharing miRNAs play a key role in the cancer development (7-9). MicroRNAs (miRNAs) are a class of typical ncRNA with approximately 22 nucleotides, which can function as a regulator to decrease the levels of mRNA (10). Long non-coding RNAs (lncRNAs) are another major kind of typical ncRNA, possessing more than 200 nucleotides, and have been regarded as miRNA sponges that reduce the levels of miRNA, thus relieving the inhibitory effect on the downstream genes that are targeted by miRNAs (11,12). Increasing experimental evidence has shown that the lncRNA-miRNA-mRNA network plays a key regulatory role in the development and progression of several cancers, such as esophageal (13), colorectal (14), gastric (15), and lung cancer (16). However, current lncRNA-miRNA-mRNA network information for human cancers is still not entirely reflective of cancer characteristics, including those of ESCA.
Although there were several similar reports about integrated analysis of CeRNA in esophageal carcinoma in PubMed (17). To the best of our knowledge, the studies that involved a large sample size with cancer grading information were not investigated thoroughly enough, and reports related to ESCA-specific RNA biomarkers or new targets of ESCA are also lacking. In this project, we explored The Cancer Genome Atlas (TCGA) database to find the specific expression genes of lncRNAs, miRNAs, and mRNAs. A series test of cluster (STC) analysis was carried out to identify a set of unique model expression tendencies. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed to predict the functions of the specific expression mRNAs that were obtained from the STC analysis. Moreover, a novel CeRNA network in ESCA was constructed to investigate the correlations among lncRNAs, miRNAs, and mRNAs. We hypothesize that all the RNAs involved in this CeRNA network may be beneficial for us to seek more promising diagnostic biomarkers or therapeutic targets for ESCA in the future. Findings of our study will be valuable for further understanding of the molecular mechanisms, disease progress, and potential treatment targets of future ESCA research.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-615).
Methods
Samples and pre-processing
The RNA-Seq and clinical characteristics data of tissue samples from patients with ESCA were downloaded from the TCGA database (https://portal.gdc.cancer.gov/, accessed on 12 September, 2019). Then, the sample data were carefully screened according to the following selection criteria: (I) patients without any other malignant tumors; (II) samples without staging information were excluded; (III) samples included lncRNA, miRNA, and mRNA detection data. Finally, a total of 142 samples (51 samples were ESCC and 91 were ECA) were retained and divided into four groups: 9 samples were paracancerous tissues used as control in subsequent analysis; 14 cancerous samples were from patients with stage I ESCA; 65 cancerous samples were from patients with stage II ESCA; 54 cancerous samples were from patients with stage III and IV ESCA. This study fully conformed to the publishing guide provided by TCGA. The sequencing data were downloaded from TCGA database, so we did not need ethics committee approval. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Screening of differentially expressed RNAs
The RNA expression profile data of ESCA from the corresponding participants were downloaded from TCGA database. The lncRNA and mRNA expression reads were normalized by TCGA RNASeqV2 system. The miRNA sequencing (miRNAseq) data of all samples that had been performed on the Illumina HiSeq 2000 (Illumina, San Diego, CA, USA) microRNA sequencing platform were also obtained from TCGA database and normalized by TCGA using default parameters. After data preprocessing, the differentially expressed analysis of RNAs containing lncRNAs, mRNAs, and miRNAs was carried out using random variance model (RVM) test method (18) in three groups respectively, namely stage I ESCA vs. control, stage II ESCA vs. control, and stage III and IV ESCA vs. control. Only the lncRNAs, miRNAs, and mRNAs with fold changes (FC) >1.5, FDR (false discovery rate) <0.05, and P<0.05 were retained in each set of analysis. Finally, the unions of differentially expressed lncRNAs, miRNAs, and mRNAs among the three groups were retained separately for further analysis. The whole workflow of this assay is shown in Figure 1.
Series test of cluster analysis for mRNAs, lncRNAs, and miRNAs
Based on the expression levels, the three unions of mRNAs, lncRNAs, and miRNAs obtained in the previous step were subjected to a series test of cluster analysis through R (version 3.5.1, Auckland, NZ). Through this analysis, we accurately and intuitively screened out the RNA clusters with significantly upregulated or downregulated trends in line with development of ESCA (Normal → Stage I → Stage II →Stage III and IV). The specific steps were performed as previously described (19,20).
GO and KEGG pathway analysis
After the abovementioned analysis, profile 2, profile 3, profile 5, profile 6, profile 11, profile 22, and profile 25 containing significantly upregulated and downregulated trend mRNAs were then analyzed. All of the mRNAs from these profiles were annotated with gene function information according to the GO database (http://geneontology.org/). Then, Fisher’s exact and multiple comparison tests were conducted to analyze the significance level of these functions with FDR <0.05 and P<0.05, so we could screen out the significant function of these mRNAs with trend of significant change. Then, all of the mRNAs from these profiles were annotated with gene signal pathway information according to the KEGG (http://www.kegg.jp/). The Fisher’s exact and multiple comparison tests were conducted to analyze the significance level of these pathways with FDR <0.05 and P<0.05, so as to screen out the significant signal pathways involved in these mRNAs.
Target prediction of miRNAs
In order to predict potential target mRNAs and lncRNAs of the miRNAs, several target gene prediction algorithms were used. We predicted potential target mRNAs of the miRNAs using miRanda (http://www.microrna.org/), Targetscan (http://www.targetscan.org/), and miRWalk (http://129.206.7.150/). After the analysis using the three algorithms, the intersection was obtained as the site of potential target mRNAs of miRNAs. Potential target lncRNAs of the miRNAs were predicted using miRanda (http://www.microrna.org/) and PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_exe.html). After analysis with these two algorithms, the intersection was obtained as potential target lncRNAs of miRNAs. On the basis of negative correlation, related pairs of miRNA-mRNA and miRNA-lncRNA were screened out for the further research.
Construction of CeRNA and protein-protein interaction networks
Combining predicted target relationship and expression levels of these RNAs, the process of constructing CeRNA network was implemented as previously described (21). Visualization of the lncRNA-miRNA-mRNA interaction network was carried out using Cytoscape software (https://cytoscape.org) (22). Afterward, in context with the co-expressed proteins corresponding to mRNAs from CeRNA network, the protein–protein interaction (PPI) network was conducted using the Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org/) in order to find the corresponding core mRNAs.
Survival analysis
Survival statistics of samples and expression data of lncRNA, miRNA, and mRNAs from the CeRNA network were analyzed by Kaplan-Meier and log-rank test to explore the relationship between the overall survival (OS) rates and gene expression levels in patients with ESCA. Then, the survival statistics and the expression levels of mRNAs, lncRNAs, or miRNAs with significant effect (P<0.05) were used to construct the corresponding survival curves.
Statistical analysis
One-way ANOVA and Student’s t test were performed with SPSS 13.0. The significance level was set at 0.05 as default to control the false discovery rate. Data were expressed as the mean ± standard deviation of at least three separate experiments. Values of P<0.05 were considered statistically significant.
Results
Cancer specific lncRNAs, miRNAs, and mRNAs in different stages of ESCA
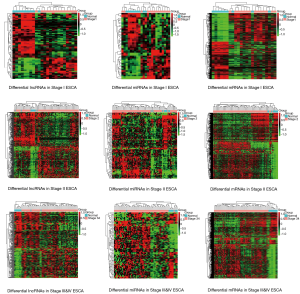
A total of 65 lncRNAs, 45 miRNAs, and 782 mRNAs were authenticated as differentially expressed between Tumor-Stage I and paracancerous tissues from TCGA database (absolute FC >1.5, P<0.05, FDR <0.05). Further, there were 333 lncRNAs, 114 miRNAs, 1990 mRNAs differentially expressed between Tumor-Stage II and paracancerous tissues (absolute FC >1.5, P<0.05, FDR <0.05). A total of 266 lncRNAs, 85 miRNAs, and 1,491 mRNAs were found to be expressed differentially between Tumor-Stage III & IV and paracancerous tissues (absolute FC >1.5, P<0.05, FDR <0.05). The number of differentially expressed lncRNAs, miRNAs, and mRNAs in all stages were 402, 125, and 2,324, respectively. All the above RNAs were used as the basis for subsequent data analysis. All of the RNAs in each independent data set were analyzed with unsupervised hierarchical clustering analysis. The heat maps respectively showed these differentially expressed lncRNAs, miRNAs, and mRNAs in all groups (Stage I ESCA vs. control, Stage II ESCA vs. control and Stage III and IV ESCA vs. control) (Figure 2).
STC analysis
In this analysis, we obtained the trends of all the RNA clusters as the progress of ESCA (Normal→ Stage I→ Stage II→ Stage III and IV). In the analysis of 2,324 mRNAs, we obtained 26 types of temporal expression pattern with the progress of ESCA. A total of 7 types (profile 2, 3, 5, 6, 11, 22, 25) marked in red were significant (P<0.05) among these types in the trend chart (Figure 3). Among these cluster profiles with significant trend, the expression of profile 22 (n=862 genes) and profile 25 (n=273 genes) showed an increased trend with the development of ESCA, while the expression of profile 2 (n=113 genes) and profile 5 (n=361 genes) revealed a trend of reduction with the development of ESCA.
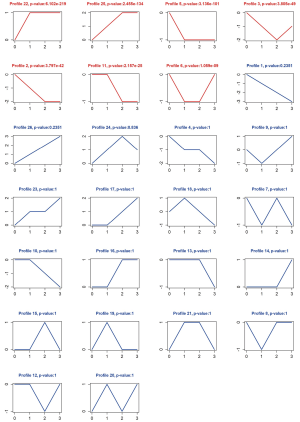
We also carried out the same STC analysis for 402 lncRNAs and 125 miRNAs. A total of 26 types of temporal expression pattern were also detected in the analysis of lncRNAs and miRNAs. In the analysis of lncRNAs, 6 types of temporal expression pattern (profile 2, 3, 5, 6, 22, 25) marked in red were significant (P<0.05) (Figure 4). Among these cluster profiles, the expression of profile 22 (n=93 genes) and profile 25 (n=81 genes) showed an increased trend with the development of ESCA, while the expression of profile 2 (n=35 genes) and profile 5 (n=99 genes) revealed a trend of reduction with the development of ESCA.

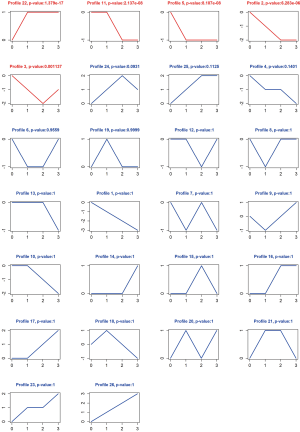
In the analysis of 125 miRNAs, 5 significant cluster profiles (P<0.05) (profile 2, 3, 5, 11, 22) among the 125 miRNAs were detected (Figure 5), and the expression of profile 22 (n=52 genes) showed an trend of increase with the development of ESCA, while the expression of profile 2 (n=10 genes) and profile 5 (n=25 genes) revealed a reduced trend with the development of ESCA.
GO and KEGG pathway analysis
All of the mRNAs from mRNA union (profile 2, 3, 5, 6, 11, 22, 25, P<0.05) that were obtained from STC analysis were analyzed to predict functions of differentially expressed genes (DEGs). The GO and KEGG pathway analyses were carried out for the upregulated and downregulated genes, respectively. Enrichment analysis has always been performed to measure the functional significance, which can help us to distinguish genes with a more optimum function feature (23).
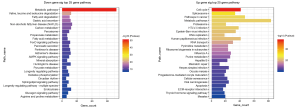
As shown in Figure 6, the upregulated genes with specific functions participate in processes of cell division, DNA replication, and negative/positive regulation of transcription from RNA polymerase II promoter, among others. These gene functions are closely related to the occurrence and development of ESCA. The downregulated genes participate in processes of negative/positive regulation of transcription from RNA polymerase II promoter, intracellular protein transport, and protein phosphorylation, among others.

The results of KEGG pathway analysis indicated that upregulated genes are related to these pathways related to cell cycle, pathways in cancer, and metabolism (Figure 7). All of these pathways were also closely related to the occurrence and development of ESCA. Downregulated genes were mainly related to various metabolic pathways.
CeRNA network construction
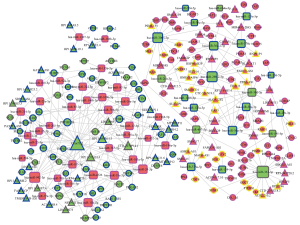
In this study, expression levels of RNAs from profiles 22, 25, 2, and 5 significantly (P<0.05) showed upregulated or downregulated trends with the development of ESCA. These four significant cluster profiles totally contained 272 lncRNAs, 87 miRNAs, and 692 mRNAs (at the same time, these mRNAs were the intersection of two significant mRNA clusters which were identified through GO and KEGG pathway analysis). Then, we predicted potential target mRNAs and lncRNAs of the miRNAs. On the basis of negative correlation, related pairs of miRNA-mRNA and miRNA-lncRNA were screened out. Finally, we constructed a lncRNA-miRNA-mRNA CeRNA network based on above related pairs for ESCA. The software Cytoscape 3.0 was used to draw a graph of the CeRNA network. At last, the CeRNA network contained 71 lncRNAs, 56 miRNAs, and 125 mRNAs (Figure 8). In the graph, the triangles represented lncRNAs, the rectangles represented miRNAs, and the balls represented mRNAs. The RNAs with larger size or shape had stronger regulatory ability in the network.
Analysis of PPIs
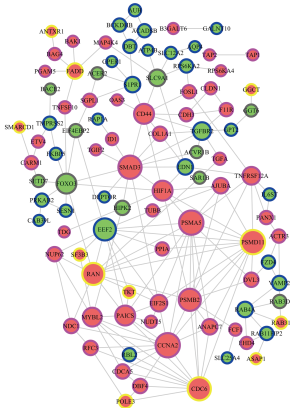
The STRING database was used to analyze the PPI network based on the mRNA that were obtained from the CeRNA network. Cytoscape 3.0 was used to draw a graph of the PPI network. Several nodes with high degrees were CD44, SMAD3, EIF2S1, RAN, FOXO3, CDC6, AJUBA, NUP62, PSMD11, HIF1A, PSMA5, PSMB2, and CCNA2, among others (Figure 9). Most of these proteins are related to the metabolism, division, and proliferation of cells.
Survival curves
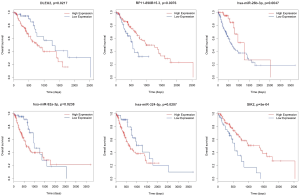
To further clarify the prognostic characteristics about the relationship between these key RNAs in the CeRNA network and the performance of ESCA patients, the OS was analyzed with univariate Cox regression analysis. We revealed the lncRNAs, miRNAs, and mRNAs significantly influenced OS (P<0.05), and then constructed the corresponding survival curve. Finally, we obtained two lncRNAs (DLEU2, RP11-890B15.3), 3 miRNAs (miR-26b-3p, miR-92a-3p, miR-324-5p), and 1 mRNA (SIK2) which significantly influenced OS (Figure 10).
Discussion
ESCA is notorious worldwide for its poor prognosis and high lethality rate. On the one hand, a lot of patients are diagnosed at an advanced stage, while on the other, there is a deficiency of efficient therapeutic targets and prognostic biomarkers; therefore, the prognosis of ESCA is very poor (1,2,5). Accordingly, focus must be given to identification of specific genes that are related to the development of ESCA, and the development of more effective diagnostic and treatment protocols for this severe disease. In the present project, we adopted TCGA database to explore the specific lncRNAs, mRNAs, and miRNAs that were most significantly associated with ESCA status. Furthermore, we attempted to clarify the specific CeRNA network in ESCA by way of lncRNA-miRNA-mRNA order pattern and identify potential prognostic RNA biomarkers. Based on comprehensive analysis, we finally identified 2 lncRNAs (DLEU2, RP11-890B15.3), 3 miRNAs (miR-26b-3p, miR-92a-3p, miR-324-5p), and 1 mRNA (SIK2) as prognostic biomarkers. High expression levels of DLEU2, miR-92a-3p, and miR-324-5p were associated with worse prognosis of ESCA, while low expression levels of RP11-890B15.3, miR-26b-3p, and SIK2 were associated with worse prognosis of ESCA.
In regards to data, lncRNAs have previously been regarded as a key oncogene and tumor suppressors with unrecognized regulation mechanisms (24). Nowadays, a volume of experimental data have shown that lncRNAs play crucial roles in biological regulatory functions, including DNA damage, epigenetic regulation, participation in signal transduction, and cell cycle regulation (25). For example, overexpressed lncRNA H19 was found to enhance the development of gastric (26), colorectal (27), breast (28), and lung cancer (29). Furthermore, many lncRNAs have been identified as potential prognostic markers and therapeutic targets for a range of different cancers.
Many studies have shown that the lncRNA DLEU2 is closely related to the occurrence and development of tumors; it has been shown to have some effects on proliferation, apoptosis, and PI3K/Akt signaling pathway of gastric cancer cells (30). The lncRNA DLEU2 promotes tumor growth by sponging miR-337-3p in human osteosarcoma (31), and it modulates the cell proliferation and invasion of non-small cell lung cancer by regulating the miR-30c-5p/SOX9 axis (32). The findings of Lu (33) revealed the pro-oncogenic role of lncRNA DLEU2 in the progression of esophageal cancer, suggesting that DLEU2 exerts ceRNA functions in ESCA through regulation of the miR-30e-5p/E2F7 axis. These studies are in good agreement with our analysis results that a high expression level of DLEU2 is associated with a worse prognosis of ESCA.
It has been clarified that the microRNAs can be detectable as stable molecules in the blood of cancer patients (34). Many studies have demonstrated that miR-26b-3p is correlated with the pathogenesis of various cancers (35,36). It has been reported that miR-26b-3p is closely related with breast cancer, intrahepatic cholangiocarcinoma, colorectal cancer (CRC), and laryngeal cancer (37). Reports have shown that miR-26b-3p is a critical modulator of glioma via its downstream molecule, ANTXR1, which indicated that the miR-26b-3p/ANTXR1 axis may be treated as a diagnostic target in glioma (38). When the expression of endogenous miR-26b-3p in glioma cells was up-regulating, the cell proliferation activity and metastasis ability increased significantly. The results of our study showed that a low-expression level of miR-26b-3p is related to the positive progress of ESCA, indicating that miR-26b-3p acts as a suppressor in ESCA.
Various studies have shown that mir-92a-3p is highly correlated with cancer. In nasopharyngeal carcinoma cells, the expression level of mir-92a-3p is related to the potential of lymph node metastasis (39). Overexpression of miR-92a-3p has been shown to significantly promote renal cell carcinoma cell proliferation and colony formation (40). The inhibition of miR-92a-3p with locked nucleic acid was shown to inhibit cell proliferation and induce apoptosis and necrosis in oncogenes in colorectal cancer (41). The miR-92a-3p is one of the differentially expressed miRNAs in the serum of ESCC patients from a high risk area (42). The miR-92a-3p promotes the proliferation, migration, and invasion of esophageal squamous cell cancer by regulating PTENb (43). These data support our findings that high expression levels of miR-92a-3p are associated with worse prognosis of ESCA.
Various research data have shown that mir-324-5p is also closely related to cancers. The migration and invasion of CRC is inhibited by mir-324-5p through targeting gene ABL2 (44). The lncRNA FOXD2-AS1 affects proliferation, migration, and invasion of colon cancer SW480 cells by targeting negatively regulated mir-324-5p expression (45). In gastric cancer cells, miR-324-5p reduces viability and induces apoptosis through modulating TSPAN8 (46). Overexpression of miR-3245p reduced the growth and invasive abilities of CRC cells (47). The study of Chiam et al. highlighted the potential of serum exosomal miR-324-5p as a biomarker for the detection of esophageal adenocarcinoma (48). Our data analysis also showed that mir-324-5p can be a valuable molecular marker in ESCA.
Salt inducible kinase 2 (SIK2) is a centrosome kinase required for mitotic spindle formation. Maxfield found that SIK2 is essential for triple-negative breast cancer (TNBC) tumor growth in vivo (49). Huang confirmed that the expression level of SIK2 protein in thyroid microcarcinoma was significantly higher than that in adjacent tissues (50). The findings of Xia indicated that SIK2 expression can serve as a prognostic biomarker for epithelial ovarian cancer (EOC) (51). Overexpression of SIK2 in ovarian cancer cells promotes abdominal metastasis while SIK2 depletion prevents metastasis in vivo (52). In our study, we showed that low expression of the SIK2 gene is related to the poor prognosis of patients with ESCA. This was also the first study to observe the relationship between the SIK2 gene and ESCA.
In the present work, we performed investigations to clarify the specific CeRNA network in ESCA by way of the lncRNA-miRNA-mRNA order pattern. Notably, based on the TCGA RNA transcript profiles collected from ESCA specimens, through CeRNA bioinformatics analysis, we identified 2 lncRNAs (DLEU2, RP11-890B15.3), 3 miRNAs (miR-26b-3p, miR-92a-3p, miR-324-5p), and 1 mRNA (SIK2). Among these RNAs, the lncRNA RP11-890B15.3 is not yet understood. Interestingly, low expression of RP11-890B15.3 was correlated with positive progress of ESCA and poor OS of the patients. Upon review of the CeRNA network we constructed, the miRNA corresponding to lncRNA RP11-890b15.3 was miR-708-5p. An important role is played by mir-708-5p in the development of gastrointestinal cancer (53), and it has been found to be a specific molecule in ESCC (54). In the CeRNA network, SIK2 was the mRNA corresponding to lncRNA RP11-890b15.3. The relationship between SIK2 and cancer has previously been repeatedly emphasized. Therefore, RP11-890b15.3 has potential value as a target in the treatment of ESCA.
In summary, we established a lncRNA-miRNA-mRNA CeRNA network in ESCA. Finally, we found 2 lncRNAs (DLEU2, RP11-890B15.3), 3 miRNAs (miR-26b-3p, miR-92a-3p, miR-324-5p), and 1 mRNA (SIK2) related to the prognosis of ESCA, which could be regarded as potential prognostic or treatment biomarkers for ESCA. Our project is significant for advancing the understanding of molecular mechanisms of ESCA development and shows a new perspective for the use of ESCA drugs and prognosis.
Acknowledgments
Funding: This study was funded by the following foundations: Key Research and Development Program Projects, Shaanxi, (2019SF-014); Bethune Charitable Foundation, (HZB-20181119-070); Shaanxi Provincial Health Research Foundation, (2018D056); Crosswise project of the First Affiliated Hospital of Xi’an Jiaotong University, (HX201739); New Clin1ical Technology of Xi’an Jiaotong University, (XJLS-2019-048); New Clinical Technology of the First Affiliated Hospital of Xi’an Jiaotong University (XJYFY-2019W3).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-615
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-615). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg 2018;41:210-5. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer Statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Chen W, Zheng R, Zeng H, et al. The incidence and mortality of major cancers in China, 2012. Chin J Cancer 2016;35:73. [Crossref] [PubMed]
- Lin Y, Totsuka Y, Shan B, et al. Esophageal cancer in high-risk areas of China: research progress and challenges. Ann Epidemiol 2017;27:215-21. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011;146:353-8. [Crossref] [PubMed]
- He XJ, Bian EB, Ma CC, et al. Long non-coding RNA SPRY4-IT1 promotes the proliferation and invasion of U251 cells through upregulation of SKA2. Oncol Lett 2018;15:3977-84. [Crossref] [PubMed]
- Luan T, Zhang XM, Wang SY, et al. Long non-coding RNA MIAT promotes breast cancer progression and functions as ceRNA to regulate DUSP7 expression by sponging miR-155-5p. Oncotarget 2017;8:76153-64. [Crossref] [PubMed]
- Xu J, Li Y, Lu J, et al. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res 2015;43:8169-82. [Crossref] [PubMed]
- Anvarnia A, Mohaddes-Gharamaleki F, Asadi M, et al. Dysregulated microRNAs in colorectal carcinogenesis: New insight to cell survival and apoptosis regulation. J Cell Physiol 2019;234:21683-93. [Crossref] [PubMed]
- Jiao D, Li Z, Zhu M, et al. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC). Am J Cancer Res 2018;8:748-60. [PubMed]
- Zhang J, Li W. Long noncoding RNA CYTOR sponges miR-195 to modulate proliferation, migration, invasion and radiosensitivity in nonsmall cell lung cancer cells. Biosci Rep 2018;38:BSR20181599. [Crossref] [PubMed]
- Tang J, Li Z, Zhu Q, et al. miR-204-5p regulates cell proliferation, invasion, and apoptosis by targeting IL-11 in esophageal squamous cell carcinoma. J Cell Physiol 2020;235:3043-55. [Crossref] [PubMed]
- Liang C, Zhao T, Li H, et al. Long Non-coding RNA ITIH4-AS1 Accelerates the Proliferation and Metastasis of Colorectal Cancer by Activating JAK/STAT3 Signaling. Mol Ther Nucleic Acids 2019;18:183-93. [Crossref] [PubMed]
- Zhu H, Dai W, Li J, et al. HOXD9 promotes the growth, invasion and metastasis of gastric cancer cells by transcriptional activation of RUFY3. J Exp Clin Cancer Res 2019;38:412. [Crossref] [PubMed]
- Ren J, Li X, Dong H, et al. miR-210-3p regulates the proliferation and apoptosis of non-small cell lung cancer cells by targeting SIN3A. Exp Ther Med 2019;18:2565-73. [Crossref] [PubMed]
- Zhao M, Wang J, Yuan M, et al. Multivariate gene expression-based survival predictor model in esophageal adenocarcinoma. Thorac Cancer 2020;11:2896-908. [Crossref] [PubMed]
- Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 2003;19:2448-55. [Crossref] [PubMed]
- Miller LD, Long PM, Wong L, et al. Optimal gene expression analysis by microarrays. Cancer Cell 2002;2:353-61. [Crossref] [PubMed]
- Ramoni MF, Sebastiani P, Kohane IS. Cluster analysis of gene expression dynamics. Proc Natl Acad Sci U S A 2002;99:9121-6. [Crossref] [PubMed]
- Li CY, Liang GY, Yao WZ, et al. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol 2016;48:1965-76. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Mutowo P, Bento AP, Dedman N, et al. A drug target slim: using gene ontology and gene ontology annotations to navigate protein-ligand target space in ChEMBL. J Biomed Semantics 2016;7:59. [Crossref] [PubMed]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391-407. [Crossref] [PubMed]
- Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer 2015;1:93-109. [Crossref] [PubMed]
- Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014;5:2318-29. [Crossref] [PubMed]
- Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015;6:22513-25. [Crossref] [PubMed]
- Si X, Zang R, Zhang E, et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 2016;7:81452-62. [Crossref] [PubMed]
- Zhang Q, Li X, Li X, et al. LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by targeting miR-484 in human lung cancer cells. J Cell Biochem 2018;119:4447-57. [Crossref] [PubMed]
- Xie HX, Xu ZY, Tang JN, et al. Effect of Huaier on the proliferation and apoptosis of human gastric cancer cells through modulation of the PI3K/AKT signaling pathway. Exp Ther Med 2015;10:1212-8. [Crossref] [PubMed]
- Liu W, Liu PC, Ma K, et al. LncRNA DLEU2 promotes tumour growth by sponging miR-337-3p in human osteosarcoma. Cell Biochem Funct 2020;38:886-94. [Crossref] [PubMed]
- Zhou Y, Shi H, Du Y, et al. lncRNA DLEU2 modulates cell proliferation and invasion of non-small cell lung cancer by regulating miR-30c-5p/SOX9 axis. Aging (Albany NY) 2019;11:7386-401. [Crossref] [PubMed]
- Lu T, Wang R, Cai H, et al. Long non-coding RNA DLEU2 promotes the progression of esophageal cancer through miR-30e-5p/E2F7 axis. Biomed Pharmacother 2020;123:109650. [Crossref] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [Crossref] [PubMed]
- Fan D, Lin X, Zhang F, et al. MicroRNA 26b promotes colorectal cancer metastasis by downregulating phosphatase and tensin homolog and wingless-type MMTV integration site family member 5A. Cancer Sci 2018;109:354-62. [Crossref] [PubMed]
- Meng C, Liu Y, Shen Y, et al. MicroRNA-26b suppresses autophagy in breast cancer cells by targeting DRAM1 mRNA, and is downregulated by irradiation. Oncol Lett 2018;15:1435-40. [PubMed]
- Tian L, Zhang J, Ren X, et al. Overexpression of miR-26b decreases the cisplatin-resistance in laryngeal cancer by targeting ATF2. Oncotarget 2017;8:79023-33. [Crossref] [PubMed]
- Geng F, Lu GF, Ji MH, et al. MicroRNA-26b-3p/ANTXR1 signaling modulates proliferation, migration, and apoptosis of glioma. Am J Transl Res 2019;11:7568-78. [PubMed]
- An JX, Ma MH, Zhang CD, et al. miR-1236-3p inhibits invasion and metastasis in gastric cancer by targeting MTA2. Cancer Cell Int 2018;18:66. [Crossref] [PubMed]
- Zeng R, Huang J, Sun Y, et al. Cell proliferation is induced in renal cell carcinoma through miR 92a 3p upregulation by targeting FBXW7. Oncol Lett 2020;19:3258-68. [Crossref] [PubMed]
- Ahmadi S, Sharifi M, Salehi R. Locked nucleic acid inhibits miR-92a-3p in human colorectal cancer, induces apoptosis and inhibits cell proliferation. Cancer Gene Ther 2016;23:199-205. [Crossref] [PubMed]
- Li SQ, Wang HM, Cao XF. Potential clinical insights into microRNAs and their target genes in esophageal carcinoma. Biomarkers 2011;16:629-36. [Crossref] [PubMed]
- Li X, Guo S, Min L, et al. miR-92a-3p promotes the proliferation, migration and invasion of esophageal squamous cell cancer by regulating PTEN. Int J Mol Med 2019;44:973-81. [Crossref] [PubMed]
- Cao L, Xie B, Yang X, et al. MiR-324-5p suppresses hepatocellular carcinoma cell invasion by counteracting ECM degradation through post-transcriptionally downregulating ETS1 and SP1. PLoS One 2015;10:e0133074. [Crossref] [PubMed]
- Peng K, Yue F, Zhang L. Effect of LncRNA FoxD2-AS1 on proliferation and migration of colon cancer SW480 cells by targeting miR-324-5p. J Clin Exp Med 2019;18:1724-8.
- Lin H, Zhou AJ, Zhang JY, et al. MiR-324-5p reduces viability and induces apoptosis in gastric cancer cells through modulating TSPAN8. J Pharm Pharmacol 2018;70:1513-20. [Crossref] [PubMed]
- Kuo WT, Yu SY, Li SC, et al. MicroRNA-324 in Human Cancer: miR-324-5p and miR-324-3p Have Distinct Biological Functions in Human Cancer. Anticancer Res 2016;36:5189-96. [Crossref] [PubMed]
- Chiam K, Wang TT, Watson DI, et al. Circulating Serum Exosomal miRNAs As Potential Biomarkers for Esophageal Adenocarcinoma. Journal of Gastrointestinal Surgery 2015;19:1208-15. [Crossref] [PubMed]
- Maxfield KE, Macion J, Vankayalapati H, et al. SIK2 Restricts Autophagic Flux To Support Triple-Negative Breast Cancer Survival. Mol Cell Biol 2016;36:3048-57. [Crossref] [PubMed]
- Huang B, Zheng JZ, Liu JH, et al. Expression and significance of sik1 and sik2 in thyroid microcarcinoma and its adjacent tissues. Medical Clinical Research 2019;36:863-6.
- Xia B, Lin M, Dong W, et al. Upregulation of miR-874-3p and miR-874-5p inhibits epithelial ovarian cancer malignancy via SIK2. J Biochem Mol Toxicol 2018;32:e22168. [Crossref] [PubMed]
- Miranda F, Mannion D, Liu S, et al. Salt-Inducible Kinase 2 Couples Ovarian Cancer Cell Metabolism with Survival at the Adipocyte-Rich Metastatic Niche. Cancer Cell 2016;30:273-89. [Crossref] [PubMed]
- Wang H, Xu T, Wu L, et al. Molecular mechanisms of MCM3AP-AS1 targeted the regulation of miR-708-5p on cell proliferation and apoptosis in gastric cancer cells. Eur Rev Med Pharmacol Sci 2020;24:2452-61. [PubMed]
- Chu Y. MicroRNA expression profiling of esophageal squamous cell induced by cigarett smoke extract and its possible mechanism. Hunan: Central South University, 2012.
(English Language Editor: J. Jones)



