
CircBACH1/let-7a-5p axis enhances the proliferation and metastasis of colorectal cancer by upregulating CREB5 expression
Introduction
In the past decade, with colorectal cancer prevention technology, the patients’ survival with early colorectal cancer has distinctly improved (1). However, the 10-year and more than 10-year survival rates of patients with advanced colorectal cancer are still exceptionally low. According to the data covering more than 30 countries, the 5-year survival rate of colorectal cancer patients is about 50–60%, and the 5-year survival rate of colorectal cancer patients in Japan and the United States is about 60–70% (2). According to the relevant data from the American Joint Committee on Cancer (AJCC), the 5-year survival rate of patients with stage I colorectal cancer at diagnosis can reach more than 90%; when colorectal cancer progresses to stage IV and V, the rate of patients decreases to 8% (3). In the past 10 years, the survival rate of colorectal cancer in China has shown a good upward trend due to the popularization of endoscopic techniques. However, compared with developed countries such as Europe, the United States, and Japan, there is still a huge gap in the survival rate. At present, the overall 5-year survival rate and long-term survival rate of colorectal cancer patients in China are low, related to the unclear molecular pathogenesis of colorectal cancer.
The incidence of colorectal cancer results from various harmful factors, which includes multiple steps with different pathophysiological stages. Precancerous lesions include adenoma and ulcerative colitis in inflammatory bowel disease. Studies on the pathogenesis of colorectal cancer reveal that there are many possible molecular pathways, and the main related genes include KRAS, β-catenin, p53, MAPK, PI3K, and c-myc (1,4-8). To improve the early diagnosis and prognosis of colorectal cancer, the molecular mechanism of its pathogenesis has always been the focus of colorectal cancer research.
With the improvement of genome sequencing, scientists have found that less than 2% of RNA in the whole genome can translate protein, and the remaining 98% cannot encode protein (called non-coding RNA). These non-coding RNAs used to be considered “noise” and had no function. Circular RNA is a closed-loop structure and can be more stable in cells. Liang et al. (9) showed that circ-ABCB10 is a tumor regulator, which can augment breast cancer cells’ proliferation and diminish the apoptosis of breast cancer cells. In non-small cell lung cancer, circMAN2B2 can augment lung cancer development by increasing the expression level of FOXK (10). Studies (11) confirmed that silencing the expression of circCCDC66 can diminish the growth, invasion, and metastasis of colon cancer. Therefore, an in-depth study of cyclic RNA’ seconds mechanism can improve our understanding of the pathogenesis of cancer and supply a new idea for improving the treatment and prognosis of patients.
In recent years, many miRNAs have been confirmed to affect tumor pathogenesis and progression by regulating mRNA. For example, Jin et al. carried out a prospective study on 455 patients who had undergone radical surgery for liver cancer (12,13). The results showed that miR-26a and miR-26b were heightened in women than in men. Compared with the corresponding noncancerous tissues, miR-26 in tumor tissues was attenuated, which showed that miR-26 was correlated with primary hepatocellular carcinoma. Also, the transcriptome pattern of the tumors with reduced miR-26 showed that the signal pathway activation between nuclear factor kappaB and interleukin-6 might play a role in tumor development. Let-7 family is the second miRNA molecule to be discovered. The downregulation of let-7a has been recognized as a key event in the occurrence and development of colorectal cancer and has been confirmed as a tumor suppressor gene. C-myc, RAS, and HMGA2 are downstream target genes of let-7. Recent studies have shown that it can regulate TGF-β1 signaling pathway and impact cell proliferation and differentiation (14). However, there is no report on the direct interaction of cyclic RNA.
The CAMP response element-binding protein 5 (CREB5) is a cAMP response element-binding protein family (15). The cAMP response element-binding 5 (CREB5) is a transcription factor of eukaryotic cells and a protein-coding gene. Its products are members of the cAMP response element (CRE) binding protein family. Earlier studies have shown that CREB5 is overexpressed in several types of human cancers. In epithelial ovarian cancer cells, CREB5 related mRNA, and protein levels were highly expressed (15). Patients with a high-level of CREB5 had shorter overall survival, while disease-free survival was negatively correlated with CREB levels. In lung cancer patients, a high level of CREB5 indicates a good disease-free survival (16). Whether CREB5 can affect colorectal cancer’ seconds biological characteristics, and if so, what is the mechanism of its action, these aspects need to be confirmed by experiments.
Hsa_ circ_ 0061395, derived from the BTB domain and CNC home 1 (BACH1) gene, enhanced the proliferation of liver cancer cells and was closely related to poor prognosis. In addition, it interacts with HuR to regulate P27 expression, thereby influencing cell cycle progression. Thus, circBACH1 can be used as a new diagnostic biomarker and therapeutic target (17). Also, in CRC, BACH1 diminishes apoptosis by regulating let-7a-5p, and CREB5 is the downstream target of let-7a-5p. BACH1 can impact the proliferation of CRC cells through CREB5. Therefore, circBACH1 can be used as a new diagnostic biomarker and therapeutic target.
We present the following article in accordance with the ARRVIE reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-498).
Methods
Patient tissue collection and information
Twenty pairs of CRC tissues and adjacent tissues were obtained from the Eastern Hepatobiliary Surgery Hospital. All patients had not received radiotherapy or chemotherapy, and all patients voluntarily signed informed consent before surgery—sample inclusion criteria: confirmed by two pathology experts independently. The clinicopathological information of the patients was abundant. No preoperative radiation, chemotherapy history. Exclusion criteria: lack of clinical information; lack of control; small tumor; insufficient sample size. The sample tissue was at once treated with liquid nitrogen and stored in the refrigerator at −80 °C. The study was approved by the Ethics Committee of the Eastern Hepatobiliary Surgery Hospital (No.: IRB-GY-2019-07) and in line with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients.
Cell culture and transfection
Cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM high glucose medium (Gibco, Life Technologies, Rockville, MA, USA) containing 10% fetal bovine serum, or in 1640 medium (Gibico, USA) containing 10% fetal bovine serum at 37 °C and 5% CO2, and all were in a logarithmic growth stage. si-NC, si-circBACH1, NC vector, vector-circBACH1, mimics-NC, let-7a-5p mimics, inhibitor-NC, let-7a-5p inhibitor, sh-NC, and sh-CREB5 were designed and synthesized by Genepharma. Then cell transfection was conducted according to the specification of Lipo2000 (Life Technologies, Rockville, MA, USA) for 48 hours.
Lentivirus infects cancer cell lines
Lentivirus expressing circBACH1 shRNA and the negative control were cloned into pLenti-EF1a-EGFP-F2A-Puro (Biovector Inc.). Briefly, the packaged plasmid pSPAX2 and pMD2G and the prepared recombinant lentivirus plasmid were transfected with Lipofectamine 2000, and the fresh complete cultured group containing 10% FBS was replaced 6 h after the staining. The screening medium was prepared by diluting puromycin with the medium by gradient, and the screening medium was changed every 2 to 3 days. The cells were cultured to logarithmic growth stage and 100 µL of lentivirus diluents were added to each well. After 72 h of infection, the percentage of fluorescent cells in each well was observed under a fluorescence microscope. At 24 h after infection, the virus-free adaptive culture medium was replaced, and the culture was continued at 37 °C for 24 h. The uninfected cells were screened by purinamycin resistance.
Flow cytometry
Cell apoptosis after different treatment was tested using the annexin V-fluorescein isothiocyanate Apoptosis Detection Kit I (BD Biosciences, San Jose, CA, USA) and assessed by fluorescence-activated cell sorting (FACS).
Transwell
After 24 hours of transfection, the cells were digested by trypsin to make a cell suspension. A mixture of Matrigel (Life Technologies, Rockville, MA, USA) and serum-free medium was added to the Transwell chamber. After the Matrigel solidified, the cell suspension was added to the Transwell upper chamber (Millipore, Billerica, MA, USA). RPMI-1640 medium holding 10% FBS was added to the lower chamber. The cells were cultured at 5% CO2 and 37 °C for 48 hours. Then Matrigel was removed with a cotton swab. The cells attached to the back of the filter membrane were fixed with 4% paraformaldehyde for 10 min and stained using 0.1% crystal violet for 30 min. Take pictures under a microscope.
Scratch test
When the confluence of the cells reached 95%, 200 µL of pipette scratches were used. After 48 hours, three visual fields were randomly selected by microscopethe and relative migration distance of the cells was determined.
Xenograft tumor model
Male nude mice aged 4–6 weeks were selected from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The LoVo cells were digested with trypsin. After centrifugation, the second supernatant was discarded, and the cells were diluted with PBS to support the cell concentration at 1×107/mL. Then cells were aspirated by syringe and inoculated into the armpit of the right forearm. Tumor volume in mice was measured daily. After three weeks, the mice were anesthetized with pentobarbital sodium and sacrificed using a CO2 release device. Tumor tissue was collected, and the tumor weight was weighed after taking photos. Experiments were performed under a project license (No. IRB-GY-2019-1012) granted by Ethics Committee of the Eastern Hepatobiliary Surgery Hospital, in compliance with Eastern Hepatobiliary Surgery Hospital guidelines for the care and use of animals.
Immunohistochemical
Tumor tissue specimens were immobilized with 10% neutral formalin. Immunohistochemical staining was performed by the SABC method in 4 µm continuous sections. All sections were routinely dewaxed, rinsed with PBS, and incubated at 3% H2O2 at room temperature for 10min to inactivate endogenous peroxidase. Citric acid is heated for antigen repair. Normal goat serum is sealed. The primary antibody was added in drops and incubated at 4 °C overnight. Then, biotinylated secondary antibodies and peroxidase-labeled antibodies were dropped and washed with PBS. DAB is used for color development and hematoxylin contrast dyeing. PBS was used as the negative control instead of the primary antibody.
Double luciferase reporter gene assay
According to TargetScan prediction of the suggested potential binding site information, the DNA fragment containing this site (WT) and the DNA fragment containing this site mutant (MUT) were synthesized. We cloned the double luciferase augmenter vector. The plasmid was co-transfected with let-7a-5p mimics—cultured for 48 hours in a 37 °C incubator with 5% CO2. The double luciferase reporter assay kit was used for detection. Wash the cells with PBS three times, add PLB lysed cells, and shake them in a shaker for 30 minutes. Cell lysis solution and LAR II mixed blowing, measuring the firefly luciferase reaction intensity. Add Stop&Glo Reagent to measure the reaction intensity of sea kidney luciferase.
RNA pull-down
The sequence of the circBACH1 probe used in this assay was 5'‐GUCACCAGCUUCUCAAUUUTT‐3'. CRC cells were transfected with 3'-biotinylated miRNA mimics at 100 nM. The RNA Pull down lysis buffer (20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 nM MgCl2, 0.3% NP-40) lysed cells. 25 µL Dynabeads MyOne avidin C1 was washed with a cleavage buffer. Rotate it at 4 degrees for 2 hours. Yeast tRNA and BSA are closed. The lysis buffer is used to wash the magnetic beads. Magnetic beads were incubated with 100 µL cell lysate and 100 µL miRNA—isolation of biotin-labeled miRNAs and their interacting RNAs. QRT-PCR was used to detect the RNA interaction of miRNA.
Western blot
Total proteins were extracted from each group of cells. Protein quantitative of BCA protein analysis kit and refer to the instructions for operation. Protein samples of 20 µg were separated by 10% SDS-PAGE and transferred to nitric acid fiber membrane by wet method. After membrane transfer, TBST containing 5% skimmed milk powder was used to seal the membrane, and then the primary anti-rabbit anti-CREB1 (1:800, Abcam, ab32515), anti-CREB3 (1:800, Abcam, ab180119), anti-CREB5 (1:800, Abcam, ab277098) and rabbit anti-GAPDH (1:800, Abcam, ab8245) were used to incubate the membrane overnight at 4 °C. After primary antibody incubation, rabbit IgG was incubated with horseradish peroxide HRP anti-rabbit IgG (1:1,000) at room temperature for 1 h, followed by color development with ECL chemiluminescence.
qRT-PCR
Total RNA was extracted from tissues or cells 24 hours after transfection using TRIzol reagent (Invitrogen) as instructed. Total RNA was purified using RNaseR reagent (Epicentre, Inc.). The Prime Script RT Reagent Kit (Takara) was used for retranscription of total RNA into cDNA. ABIVII 7 real-time fluorescent quantitative PCR (BIO-RAD) was used to detect the target gene’s relative expression levels in tissues and cells. The specific primer sequences used in this study are in Table 1. The reaction conditions were as follows: 95 °C 10 minutes, 95 °C 15 seconds, 60 °C 1 minutes (40 cycles). GAPDH is used as an inteRNAl reference gene. The 2−ΔΔCt method of relative gene level.
Full table
CCK-8 assay
After transfection, cell activity was evaluated with CCK-8 reagent (Beijing Solebo Technology Co., Ltd.). Cells were digested with a 0.25% trypsin solution to produce cell suspension. Cell suspension (100 µL/well) was inoculated in 96-well plates, and the plates were incubated at 37 °C containing 5% CO2 for 48 hours. 10 µL CCK-8 reagent was added to each well. The cells were cultured at 5% CO2 and 37 °C for 2 hours. The cells’ absorbance was measured at 450 nm with a microplate reader (MultiskanEX, Lab systems, Helsinki, Finland).
Statistical analysis
SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis, and the measurement data were as mean ± standard deviation. One-way ANOVA was used for comparison between. A t-test was used to compare the twos—P<0.05 means that the difference was statistically significant.
Results
Overexpression of circRNA BACH1 in CRC tissues and cells
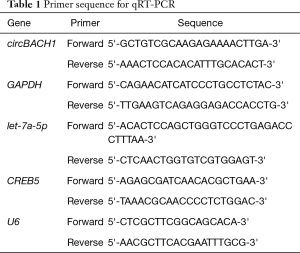
BACH1 levels in CRC and adjacent tissues were evaluated in the collected clinical samples. The level of BACH1 in CRC was distinctly enhanced than that in the adjacent tissues (P<0.01, Figure 1A) with statistical significance. QRT-PCR evaluated the BACH1 level in colorectal cancer cell line and normal colorectal cell line FHC. The results displayed that BACH1 was overexpressed in colorectal cancer cells, of which the highest level was found in the LoVo cell line (P<0.01, Figure 1B). Therefore, the LoVo cell line was selected for later experiments.
The relationship between let-7a-5p and BACH1
The level of BACH1 was meaningfully attenuated in LoVo cells, which were transfected with si-BACH1 (P<0.01, Figure 1C). By searching the database, we found that there is some correlation between let-7a-5p, miR-98-5p, miR-196a-5p and miR-155-5p, and BACH1. The results proved that miRNA was distinctly enhanced than control (P<0.05, Figure 1D), and let-7a-5p was most significant.
Therefore, we chose let-7a-5p as the researchers aim to conduct follow-up experiments. Compared with the adjacent tissues, let-7a-5p level in CRC was distinctly attenuated (P<0.01, Figure 1E) and BACH1 may target let-7a-5p (Figure 1F). Furthermore, BACH1 was meaningfully heightened than that of the matched (P<0.01, Figure 1G). The addition of let-7a-5p mimics can distinctly diminish the luciferase activity of the CRC cells transfected with BACH1-wt, while the CRC cells transfected with BACH1-mut are not impacted (Figure 1H). To further evaluate the interaction between BACH1 and let-7a-5p at the transcription level, experiments were implemented to decipherd that the BACH1 level in the let-7a-5p mimics was not altered (P>0.05, Figure 1I). The results show that the two are a one-way adjustment. Moreover, let-7a-5p had a negative correlation with the BACH1 level (Figure 1J). Collectively, we conclude that let-7a-5p is the target of BACH1 in CRC cells.
BACH1 regulates the influence of let-7a-5p on the proliferation, metastasis, and invasion of CRC cells
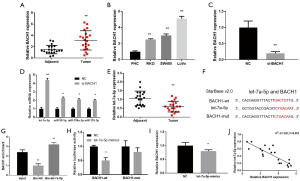
QRT-PCR revealed that let-7a-5p was distinctly accelerated in the let-7a-5p mimics (P<0.01, Figure 2A). Correspondingly, in the let-7a-5p inhibitor, let-7a-5p level attenuated distinctly (Figure 2A). The proliferation potential of CRC cells transfected with let-7a-5p inhibitor was distinctly heightened than that of the control (Figure 2B). The proliferation potential of CRC cells with let-7a-5p mimics or si-BACH1 was meaningfully lower (Figure 2B). However, the proliferation of CRC cells with si-BACH1 + let-7a-5p inhibitor was not distinctly different from that of the control. The Transwell asset and Scratch test measured the invasion and migration potential of CRC cells. The invasion and migration potential of CRC cells were meaningfully enhanced after let-7a-5p inhibitor (P<0.01, Figure 2C,D,E), and then attenuated after let-7a-5p mimics or si-BACH1 (P<0.01, Figure 2C,D,E). The transfection of si-BACH1 + let-7a-5p inhibitor was not distinctly different. Our results show that BACH1 augments cell malignancy by regulating let-7a-5p in CRC cells.
The influence of BACH1 on apoptosis through let-7a-5p
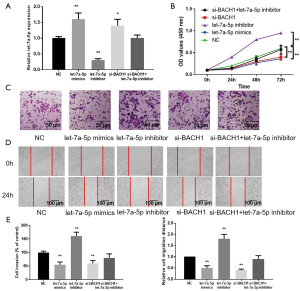
Flow cytometry assay was implemented to measure apoptosis. The apoptosis rate revealed that the apoptosis rate of let-7a-5p and si-BACH1 accelerated distinctly (Figure 3A,B). While let-7a-5p inhibitor was meaningfully attenuated (Figure 3A,B) and si-BACH1 + let-7a-5p inhibitor was similar to that of control (P>005, Figure 3A,B). Our results show that BACH1 can reduce the apoptosis of CRC cells through let-7a-5p.
The in vitro tests were used to detect the level of CREB5 in colon cancer and adjacent tissues, which supported the above conclusions (Figure 3C,D).
Relationship between let-7a-5p and CREB5
Targetscan 7.1 predicts that let-7a-5p may target CREB5 (Figure 4A). The addition of let-7a-5p mimics can inhibit the luciferase activity of Lovo cells with wild CREB5, but not CREB5-mut, suggesting that let-7a-5p can directly diminish the level of CREB5 in combination with the three non-coding regions of CREB5 (Figure 4B). In LoVo cells transfected with let-7a-5p inhibitor and mimics, CREB5 in mRNA and protein levels was measured. Western blot and QRT-PCR were implemented to confirm that let-7a-5p may diminish the level of CREB5. Compared with the control, the expression of let-7a-5p inhibitor accelerated (Figure 4C), and let-7a-5p mimics attenuated. Let-7a-5p was negatively correlated with the level of CREB5 (Figure 4D). Also, in terms of CREB5 protein change, there is a similar trend as mRNA (Figure 4E,F). Based on the above experimental results, it is concluded that CREB5 is the downstream target of let-7a-5p in CRC cells.
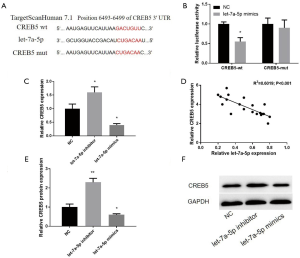
The influence of let-7a-5p on CRC cells by regulation of CREB5
qRT-PCR results revealed that CREB5 enhanced the level of CREB5 successfully to confirm targeting influence on let-7a-5p on CREB5 in CRC (P<0.01, Figure 5A). Si-CREB5 attenuated the CREB5 level efficiently. Of note, CREB5 + let-7a-5p accelerated the level of CREB5 in CRC. Transfection of si-CREB5 suppressed the proliferation of CRC cells while the proliferation potential of CRC cells with CREB5 + let-7a-5p mimics was like that of the control (Figure 5B). Transwell migration assay and wound healing assay revealed comparable results, as shown in Figure 5C,D. The results of flow cytometry revealed that the apoptosis rate of CREB5 was meaningfully attenuated while si-CREB5 was distinctly accelerated. There was no significant difference between control and CREB5 + let-7a-5p mimics (Figure 5E). In addition, Transwell migration assay and wound healing assay revealed that let-7a-5p significantly increased the migration distance, whereas the presence of pcDNA-CREB5 only partially attenuated this effect by let-7a-5p (P<0.01, Figure S1). These results show that let-7a-5p suppresses CRC cell malignancy and augments CRC cell apoptosis by targeting CREB5.
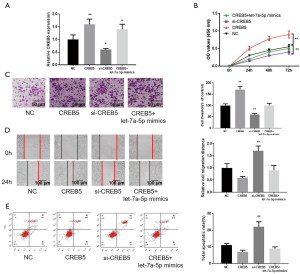
In vivo experiments confirmed the role of BACH1 in the formation of CRC
The experiment was divided into si-BACH1 and control (NC) group. LoVo cells were implanted subcutaneously in BALB/nude mice. si-BACH1 distinctly attenuated the growth and volume of subcutaneous tumors in mice (Figure 6A,B). BALB/C nude mice were killed on the 25th day, and the subcutaneous tumor was removed. In LoVo cells transfected with si-BACH1, the tumor mass was distinctly smaller (Figure 6C). Western blot revealed that CREB was distinctly suppressed in LoVo cells transfected with si-BACH1 (P<0.01, Figure 6D). In conclusion, BACH1 can affect the proliferation of CRC by regulating CREB5 in vivo.

Effects of circRNA BACH1 and let-7a-5p on CREB protein family in CRC cells
Subsequently, we confirmed the influence of on circRNA BACH1 and let-7a-5p on the CREB protein family level. The results revealed that the protein levels of CREB1 and CREB3 in si-BACH1 were lower than those in control (Figure 7A). When treated with a let-7a-5p suppressor or let-7a-5p mimics, CREB1 and CREB3 proteins’ change trend was like that of CREB5 (Figure 7B).
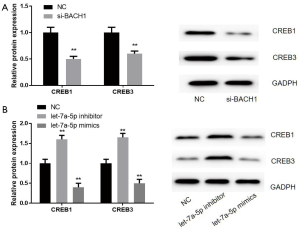
Discussion
Circular RNA (cirRNA) is a new type of non-coding RNA (ncRNA), which has the characteristics of universality, conservation, tissue specificity, and rich biological functions, and participates in various physiological and pathological processes. The expression of circRNA was significant in various tumor tissues and adjacent normal tissues. It can be used as a new biomarker and therapeutic target. It has been reported that there are a series of abnormally expressed circRNAs in colorectal cancer (CRC), including ciRS-7 (18), hsa-circ 0000069 (19), circ-HIPK3 (20) and hsa-circ000984 (21), which are up-regulated and play the role of an oncogene in CRC. Most of these circRNAs impact the proliferation, invasion, and metastasis of tumor cells through the mechanism of competitive endogenous RNA (ceRNA) and are closely related to the clinicopathological factors of CRC patients.
In this study, the expression of BACH1 in CRC was meaningfully enhanced than that in FHC normal mucosa. We confirmed that the expression of BACH1 in CRC was distinctly up-regulated by QRT-PCR, which was consistent with earlier studies. Simultaneously, in this project, we compared different kinds of colorectal cancer cell lines and chose the LoVo cell line according to the expression level. Through the scratch test and the Transwell test, we confirmed that overexpression of BACH1 could augment the metastasis and migration of CRC. Through CCK8 and flow cytometry, we confirmed that BACH1 could reduce the apoptosis and augment the proliferation of CRC cells.
MiRNA not only plays an epigenetic role in the development of viral hepatitis, liver cirrhosis, and liver cancer but also regulates the occurrence and development of colorectal cancer. It has been suggested that down-regulation of AP4 expression by miR-15a essential to induce mesenchymal-epithelial transformation (MET) and cell cycle arrest (22). Also, miRNA-29b, let-7, miRNA-141, and other miRNAs can augment EMT by diminishing ZEB/H-ras/HMGA2/MMP/snail and other target genes and ultimately impact the progress of colorectal cancer (23-25). Let-7 is a miRNA superfamily with 12 members, which was first discovered in the study of Caenorhabditis elegans (14). A large number of studies have reported that let-7 can act as a tumor suppressor gene in a variety of malignant tumors, impacting the occurrence and development of tumors. Let-7 is also reported to be involved in the epithelial mesenchymal transition of tumor cells (26). Peter et al. showed that ARBA distinctly upregulated let-7 and miR-200 family in various CRC cell lines, and the latter could augment epithelial mesenchymal transition of CRC cells by regulating the expression of Cdk6, vimentin and E-cadherin (27). The cells were transfected with si-BACH1, let-7a-5p mimics and let-7a-5p inhibitor, then qRT-PCR and biotinylated let-7a-5p was used. The cells were transfected with let-7a-5p mimics, let-7a-5p inhibitor, si-BACH1, si-BACH1 + let-7a-5p CCK-8, and Transwell and wound repair experiments confirmed that BACH1 augmented the proliferation, migration and metastasis of CRC by regulating let-7a-5p.
Studies on the function of CREBs have shown that knockout of the CREB1/CREB5 gene can increase tumor necrosis factor-alpha (TNF-α) and enhance the level of Phospho-NFkB in human monocytes (28). In our current experiment, the level of CREB5 in colorectal cancer cells accelerated meaningfully; the increase of the CREB5 level can augment the proliferation, metastasis, and migration and attenuate the apoptosis of colorectal cancer cells. The mechanism of colorectal cancer-related circRNA regulating this gene is diverse, including the following aspects: epigenetic modification, the interaction between circRNA and miRNA, interaction between circRNA and protein, and acting as a precursor or pseudogene of mRNA.
After a series of experiments, we confirmed the key role of circRNA BACH1 in CRC tumorigenesis and elucidated the interaction between BACH1, let-7a-5p, and CREB5. Through our experiments, we concluded that: in colorectal cancer cells, let-7a-5p is the downstream target of CREB5, and CREB5 is the downstream target of let-7a-5p. It is concluded that circRNA BACH1 controls CRC progression by targeting let-7a-5p and CREB5.
Overall, our study confirmed that the levels of BACH1 and CREB5 accelerated, while the expression of let-7a-5p was lowered in colorectal cancer cells. These different expressions enhance the proliferation, metastasis, and migration of colorectal cancer cells and attenuate colorectal cancer cells’ apoptosis. Finally, we draw the following conclusion that BACH1 regulates CREB5 by targeting let-7a-5p, enhances the proliferation, metastasis, and migration of colorectal cancer cells, and attenuates cell apoptosis. Our studies have proved the molecular mechanism of circRNA BACH1 acting on colorectal cancer. This molecular mechanism can be used as a therapeutic target to augment the treatment of colorectal cancer.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-498
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-498
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-498). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of the Eastern Hepatobiliary Surgery Hospital (No. IRB-GY-2019-07) and in line with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients. Animal experiments were performed under a project license (No. IRB-GY-2019-1012) granted by Ethics Committee of the Eastern Hepatobiliary Surgery Hospital, in compliance with Eastern Hepatobiliary Surgery Hospital guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Parkin DM, Bray F, Ferlay J, et al. Global Cancer Statistics, 2002. CA Cancer J Clin 2005;55:74-108. [Crossref] [PubMed]
- Gbolahan O, O'Neil B. Update on systemic therapy for colorectal cancer: biologics take sides. Transl Gastroenterol Hepatol 2019;4:9. [Crossref] [PubMed]
- Zhang H, Song J, Ren H, et al. Detection of low-abundance KRAS mutations in colorectal cancer using microfluidic capillary electrophoresis-based restriction fragment length polymorphism method with optimized assay conditions. PLoS One 2013;8:e54510. [Crossref] [PubMed]
- Murad NAA, Othman Z, Khalid M, et al. Missense mutations in MLH1, MSH2, KRAS, and APC genes in colorectal cancer patients in Malaysia. Dig Dis Sci 2012;57:2863-72. [Crossref] [PubMed]
- Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol 2005;6:322-7. [Crossref] [PubMed]
- Huang Q, Shen Z, Zang R, et al. Identification of novel genes and pathways in colorectal cancer exosomes: a bioinformatics study. Transl Cancer Res 2018;7:651-8. [Crossref]
- Sharrard RM, Royds JA, Rogers S, et al. Patterns of methylation of the c-myc gene in human colorectal cancer progression. Br J Cancer 1992;65:667-72. [Crossref] [PubMed]
- Liang HF, Zhang XZ, Liu BG, et al. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res 2017;7:1566-76. [PubMed]
- Ma X, Yang X, Bao W, et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem Biophys Res Commun 2018;498:1009-15. [Crossref] [PubMed]
- Xiang D, Li Y, Lin Y. Circular RNA circCCDC66 Contributes to Malignant Phenotype of Osteosarcoma by Sponging miR-338-3p to Upregulate the Expression of PTP1B. Biomed Res Int 2020;2020:4637109. [Crossref] [PubMed]
- Jin F, Wang Y, Li M, et al. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis 2017;8:e2540. [Crossref] [PubMed]
- Liu F, Gao XJ, Lu X, et al. Progress on the relationship between miR-26 family members and malignant tumors. Chinese Bulletin of Life Sciences 2014;26:1222-8.
- Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901-6. [Crossref] [PubMed]
- He S, Deng Y, Liao Y, et al. CREB5 promotes tumor cell invasion and correlates with poor prognosis in epithelial ovarian cancer. Oncol Lett 2017;14:8156-61. [Crossref] [PubMed]
- Xia Y, Zhan C, Feng M, et al. Targeting CREB pathway suppresses Small Cell lung Cancer. Mol Cancer Res 2018;16:825-32. [Crossref] [PubMed]
- Sun J, Hoshino H, Takaku K, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase‐1 gene. EMBO J 2002;21:5216-24. [Crossref] [PubMed]
- Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7—A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res 2017;23:3918-28. [Crossref] [PubMed]
- Guo JN, Li J, Zhu CL, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther 2016;9:7451-8. [Crossref] [PubMed]
- Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 2018;9:417. [Crossref] [PubMed]
- Xu XW, Zheng BA, Hu ZM, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget 2017;8:91674-83. [Crossref] [PubMed]
- Shi L, Jackstadt R, Siemens H, et al. p53-induced miR-15a/16-1 and AP4 form a double-negative feedback loop to regulate epithelial-mesenchymal transition and metastasis in colorectal cancer. Cancer Res 2014;74:532-42. [Crossref] [PubMed]
- Smits KM, Paranjape T, Nallur S, et al. A Let-7 MicroRNA SNP in the KRAS 3′UTR Is Prognostic in Early-Stage Colorectal Cancer. Clin Cancer Res 2011;17:7723. [Crossref] [PubMed]
- Hu M, Xia MG, Chen X, et al. MicroRNA-141 regulates Smad interacting protein 1 (SIP1) and inhibits migration and invasion of colorectal cancer cells. Dig Dis Sci 2010;55:2365-72. [Crossref] [PubMed]
- Li L, Guo Y, Chen Y, et al. The Diagnostic Efficacy and Biological Effects of microRNA-29b for Colon Cancer. Technol Cancer Res Treat 2016;15:772-9. [Crossref] [PubMed]
- Liang T, Han L, Guo LJC, et al. Rewired functional regulatory networks among miRNA isoforms (isomiRs) from let-7 and miR-10 gene families in cancer. Comput Struct Biotechnol J 2020;18:1238-48. [Crossref] [PubMed]
- Peter ME. Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression. Cell Cycle 2009;8:843-52. [Crossref] [PubMed]
- Gerritsen ME, Neish AS, Moore S, et al. CREB-binding protein/p300 are transcriptional coactivatorsofp65. Proc Natl Acad Sci U S A 1997;94:2927-32. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)


