
Circulating tumor DNA as a potential prognostic and predictive biomarker during interventional therapy of unresectable primary liver cancer
Introduction
Primary liver cancer (PLC) is the six most common cancer and the second most frequent cause of worldwide cancer-related death (1,2). Hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC) occupy more than 90% of PLC. Most PLC patients were diagnosed when already in the advanced stage (3,4). Surgical resection and liver transplantation are potentially curative treatments in the early stage, while interventional therapy is the main palliative treatment for advanced patients (5). Target therapy and immunotherapy will also be used for advanced patients. Natural compounds and nanotechnology may provide better outcomes with lower systemic toxicity and fewer side effects for liver cancer patients. Currently, computed tomography (CT) and magnetic resonance imaging (MRI) are recommended as the mainly diagnosis and surveillance methods for patients with advanced liver cancer. Accuracy of both methods may be challenged by the drug perfusion during interventional therapy.
Study of markers that can be used to predict the success of molecular-targeted therapies. Prognostic markers are tended to be considered have an association with some clinical outcomes and may be considered in the clinical management of a patient (6). Predictive markers are generally used to make some specific choices between treatment options (6). Alpha fetoprotein (AFP) is often used in auxiliary diagnosis and therapeutic evaluation, but limited in insufficient sensitivity and specificity for surveillance (7,8). Therefore, there is an unmet clinical need of a biomarker with more accurate in evaluating efficiency of interventional therapy.
Liquid biopsy, especially circulating cell-free tumor DNA (ctDNA) from plasma had a high sensitivity and specificity in cancer detection at early stage, showed potential values in tumor size assessment of liver cancer (9). ctDNA was also reported to be associated with clinicopathologic parameters of liver cancer (10-12). In the previous studies, the liver cancer-associated chip region was performed to evaluate ctDNA (13,14). However, the Nest-generation sequencing had begun to become the standard for evaluating ctDNA in generally cancer species. However, the clinical value of ctDNA in prognosis or treatment effect is still unclear.
In this study, we aimed to reveal the clinical value of ctDNA in prognosis or treatment effect of interventional therapy in patients with advanced liver cancer. We screened mutations of the 1,021 cancer-related genes by sequencing from pre-treatment and in-treatment blood samples over time, and analyzed the relation between ctDNA abundance and clinical characteristics, prognosis and therapeutic evaluation. Pretreatment ctDNA was also analyzed to explore its feasibility to replace biopsy in calculated TMB for immune-therapy decision. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-409).
Methods
Patients and samples preparation
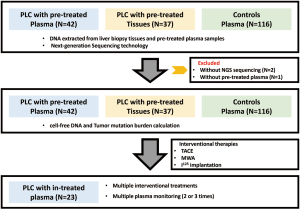
From December 2016 to April 2018, forty-five patients diagnosed with PLC at Zhuhai People’s Hospital and Shenzhen People’s Hospital, China, were enrolled in this study. The analyzed cohort included 43 HCC and 2 ICC patients (Figure S1). Three patients were excluded for non-sufficient plasma for sequencing (Figure S1). All patients provided written informed consent and received interventional therapies, the study was approved by the Ethic Committee at Zhuhai and Shenzhen Hospital, Jinan University (Table 1, the supplementary table at http://fp.amegroups.cn/cms/f29d0157930ad8f2a5b1ad554e603852/JGO-20-409-1.xlsx). Tumor burden was measured to evaluate clinical response according to the modified Response Evaluation Criteria in Solid Tumors (mRECIST 1.1). Tumor size was evaluated by the maximum tumor diameter. The Pre-treatment and in-treatment peripheral blood (10 mL) were sampled. Matched biopsy tissues were sampled when available. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).

Full table
Peripheral blood was collected in Streck tubes (Streck, Omaha, NE, USA) and processed within 72 h to separate plasma and buffy coat (source of germline DNA).
DNA extraction
Plasma was separated by centrifugation at 1600 g for 10 min, transferred to new microcentrifuge tubes, and centrifuged at 16,000 g for 10 min to remove remaining cell debris. Peripheral blood lymphocytes (PBLs) from the first centrifugation were used for the extraction of germline genomic DNA. PBL DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Circulating DNA was isolated from plasma using a QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). Buffy coat and tumor tissue DNA were extracted using the DNeasy Blood & Tissue Kit (Qiagen). DNA concentration was measured using a Qubit fluorometer and the Qubit dsDNA HS (High Sensitivity) Assay Kit (Invitrogen, Carlsbad, CA, USA). The size distribution of the cfDNA was assessed using an Agilent 2100 BioAnalyzer and a DNA HS kit (Agilent Technologies, Santa Clara, CA, USA).
Targeted capture sequencing
Before library construction, 1 µg of each tissue or buffy coat DNA was sheared to 300 bp fragments with a Covaris S2 Ultrasonicator. Indexed Illumina NGS libraries were prepared from tissue, buffy coat and circulating DNA using the KAPA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA) as previously described (15). Libraries were hybridized to custom-designed biotinylated oligonucleotide probes (Integrated DNA Technologies, Iowa, IA, USA). Capture probe was designed to cover coding sequencing or hot exons of 1,021 genes frequently mutated in solid tumors. Genes and coordinated of selected regions of each version are provided in http://fp.amegroups.cn/cms/021a5acaa5b56a29beb10078970365fa/JGO-20-409-2.xlsx, http://fp.amegroups.cn/cms/65d55f16e43d3c178de57cf9a9b5bdab/JGO-20-409-3.xlsx. Sequencing was carried out using Illumina 2×100 bp paired-end reads on an Illumina HiSeq 3000 instrument according to the manufacturer’s recommendations using a TruSeq PE Cluster Generation Kit v3 and a TruSeq SBS Kit v3 (Illumina, San Diego, CA, USA). Hybridization capture sequencing revealed a mean effective depth of coverage of 1,831× in plasma and 890× in tissue samples (http://fp.amegroups.cn/cms/41d87a1fd1be91689946c03d9513eb31/JGO-20-409-4.xlsx).
Sequence data analysis
Terminal adaptor sequences and low-quality reads were removed from raw data of paired samples. Burrows-Wheeler Aligner (BWA, version 0.7.12-r1039) tool used to align clean reads to the reference human genome (hg19). Somatic mutations were detected in circulating and tissue DNA. Non-synonymous mutations including SNVs or InDels using MuTect (version 1.1.4) and GATK, respectively, and hotspot variants was reviewed by NChot software. Clonal hematopoietic mutation was filtered as previously described, including those in DNMT3A, IDH1, and IDH2 and specific alterations within ATM, GNAS or JAK2 (16,17). The final candidate variants were all manually verified in the Integrative Genomics Viewer.
Tumor mutation burden (TMB) calculation
TMB is calculated from the pan-cancer panel. Tissue TMB (tTMB) analysis interrogated SNVs and small Indels with the variant allele frequency ≥3% (18). Blood TMB (bTMB) analysis interrogated SNVs and small Indels with all the mutations if one gene variant allele frequency ≥0.5% (18). TMB-high (TMB-H) patients were identified with 7.2 mutations/MB in liver cancer in geneplus database, and others were regarded as TMB-low (TMB-L) cohort.
Follow up
The study censored on December 30, 2019. A standardized follow-up protocol was adopted for all patients. The patients attended follow-up visits with computed tomography or abdominal magnetic resonance imaging scans about every 3 months. PFS was defined as the interval between of first interventional therapy and the date of patients had the progression disease or the latest imaging test. OS was defined as the interval between the date of first interventional therapy and the date of patients’ death or the last follow-up visit.
Statistical analysis
Pearson correlation was used to test the linear association analysis. Nonparametric comparisons were made using Wilcoxon t test. Univariate comparisons of proportion were made using a Fisher’s exact test. All statistical analyses were performed with SPSS (v.21.0; STATA, College Station, TX, USA) or GraphPad Prism (GraphPad Software 7.0, La Jolla, CA, USA) software. Statistical significance was defined as a two-sided P value of <0.05.
Results
Trails profile, patients and clinical features
In this study, 42 patients enrolled in this trial eligibly (Figure S1). Baseline characteristics were depicted in Table 1, the supplementary table at http://fp.amegroups.cn/cms/f29d0157930ad8f2a5b1ad554e603852/JGO-20-409-1.xlsx. Enrolled patients included 33 males and 9 females. The median age was 60.5 years; 12 patients involved in BCLC C stage; 66.7% of the patients were infected with hepatitis B virus (HBV) and developed cirrhosis. Hepatic ascites was occurred in 8 patients, while 25% of them (2/8) did not have HBV infection. Multifocal lesions were occurred in 50% (21/42) of the cases. The largest tumor diameter ranged from 17–200 mm, 75 mm in median. About 28.6% patients (12/42) had portal vein thrombosis (PVTT) occurred, and all of them belonged to C stage BCLC standard. The level of AFP ranged from 1.3 to 715,467 ng/mL, with a median of 43.71 ng/mL.
Mutation profiling from pre-treatment cell-free DNA
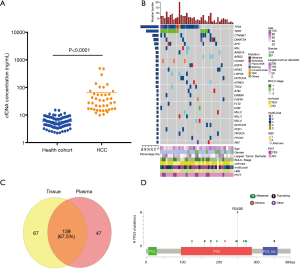
We first analyzed circulating DNA extracted from pretreatment samples and compared with that from healthy cohort (116 subjects). The median of cfDNA concentration of HCC was 28.2 ng/mL plasma (ranged from 9.8 to 487.1 ng/mL plasma), significantly higher than that of a healthy cohort (median of 5.3 ng/mL plasma, ranged from 2.2–15.1 ng/mL plasma, P<0.0001) (Figure 1A; the supplementary table at http://fp.amegroups.cn/cms/8107d522b91a4c4fc449393ac9a71bcf/JGO-20-409-5.xlsx). Based on targeted capture sequencing, a total of 187 mutations (InDels or SNVs) were detected in 40 plasma samples, with a positive rate of 95.24% (40/42, Figure 1B & http://fp.amegroups.cn/cms/46e355186179508b2a0a4e10ad2bed67/JGO-20-409-6.xlsx). In order to validate these mutations, we subsequently sequenced 37 matched tissue samples (http://fp.amegroups.cn/cms/11f672ab7d8d1c2f304a5207aa6c800d/JGO-20-409-7.xlsx). In total, 139 tumor-derived mutations were detected in plasma samples, with a consistency of 67.5% (Figure 1C). Further analysis showed consistent mutations in both tissue and plasma presented a higher mean variant allele frequency (mean VAF =23.5%) in tissue samples than mutations detected only in tissues (mean VAF =6.3%). This result suggested clonal mutation were detected more feasibly in pretreatment plasma than subclonal mutation. In ctDNA, the most frequently mutant genes were TP53 (52.4%), TERT (35.7%) and CTNNB1 (16.7%), which was consistent with previous studies (Figure 1B; http://fp.amegroups.cn/cms/46e355186179508b2a0a4e10ad2bed67/JGO-20-409-6.xlsx) (19). Mutations in TP53 including R249S, H193R, G266V, etc., among which R249S occurred with the highest mutation frequency (38.1%) (Figure 1D). Mutations in TERT occurred almost in promoter regions (91.7%).
ctDNA abundance as a more accurate biomarker than AFP in predicting tumor size
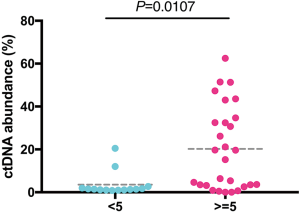
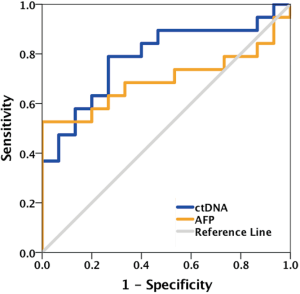
Next, we analyzed the ctDNA abundance with the max VAF of detected mutations, and evaluated the relation of ctDNA abundance, tumor size of primary cancer and AFP. The median of ctDNA abundance was 3% (range from 0 to 62.5%). Coefficient of variation (CV) was 1.2 in ctDNA abundance and lower than that of AFP (3.6). To compare the two biomarkers in reflecting the tumor size, we further evaluate their relation with tumor size. ctDNA showed a better correlation (Pearson r=0.7, P<0.0001) with tumor size, than AFP (Pearson r=0.4, P=0.0091) (Figure 2A,B). A positive correlation was identified between ctDNA abundance and AFP. Patients with ≥400 ng/mL AFP concentration had much higher ctDNA abundance than those <400 ng/mL AFP (P=0.0134) (Figure 2C). Also, we obtained a significant higher ctDNA abundance in ≥5 cm tumor size, which were considered as an important index in staging of liver cancer (Figure S2). ROC analysis also showed ctDNA abundance was more accurate biomarker than AFP in predicting tumor size, especially in assessed tumor size with 3.5 cm, with AUC of 0.82, sensitivity of 0.75 and specificity of 1 (Figures 2D,S3).

ctDNA abundance could be used to evaluate prognosis
BCLC stage, baseline AFP level, tumor dimeter, which are extension reported to assess prognosis of liver cancer (20,21). Univariate analyses were performed on age, gender, or clinicopathologic variables to determine their associations with PFS and OS (Tables 2 and 3). BCLC stage and tumor diameter could be used to predicted patients’ prognosis in our study. For advanced patients, BCLC stage and tumor diameter still had some limitations.
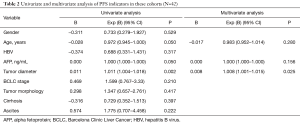
Full table
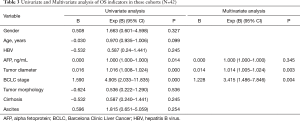
Full table
In our study, we found patients with ctDNA abundance in BCLC C stage patients was much higher (median =25%) than A and B stage (median =2%, P=0.0005) (Figure 3A). During interventional therapy, 9 patients suffered progressed disease, and others were under disease control at the first therapy evaluation. PD patients were enriched in TP53 mutation group compared with TP53 wildtype group (P=0.0221), indicated TP53 mutation may be a potential factor of poor prognosis in advanced liver cancer (Figure 3B,C). Ten patients showed no progression or had censored and 20 patients had been died in the follow-up time. TP53 wildtype group had a longer first-line PFS (P=0.411), and the median PFS time was 3 months in TP53 mutation group and 4.5 months in TP53 wildtype group (Figure 3D). 63.6% (14/22) TP53 mutation patients had been died in 35 months while only 30% TP53 wildtype patients had been died (P=0.0365). The median survival time was 12.5 months in TP53 mutation group and patients in TP53 wildtype group had not reach the median survival time (P=0.0589) (Figure 3E,F).
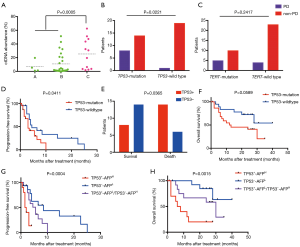
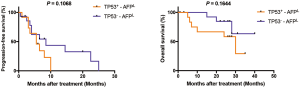
The usage of AFP in the follow-up of advanced patients still had some limitations, especially we found that 74.1% (20/27) patients with baseline AFP <400 ng/mL still had relapsed within a short period (median PFS time was 6 months). We classified patients with baseline AFP <400 ng/mL into TP53 mutation group (TP53+-AFPL) and TP53 wildtype group (TP53−-AFPL), TP53−-AFPL patients had a better PFS and OS curve than TP53+-AFPL group, although there was no statistical difference (Figure S4). The median PFS time was 5.5 months in TP53 mutation group and 8.5 months in TP53 wildtype group, and the median overall survival time was 30 months in TP53 mutation group and TP53 wildtype group had not reach the median survival time (Figure S4). Combining TP53 and AFP, we divided all patients into three groups, TP53 mutation and AFP ≥400 ng/mL (TP53+-AFPH), TP53 mutation and AFP <400 ng/mL or TP53 wildtype and AFP ≥400 ng/mL (TP53+-AFPL/TP53−-AFPH) and TP53 wildtype and AFP <400 ng/mL (TP53−-AFPL). TP53−-AFPL group patients had the better PFS (P=0.0004) and OS (P=0.0015) (Figure 3G,H). The median PFS was 1.75, 5.5 and 8.5 months in TP53+-AFPH, TP53+-AFPL/TP53−-AFPH, and TP53−-AFPL group, respectively. The median OS was 7 and 30 months in TP53+-AFPH and TP53+-AFPL/TP53−-AFPH group, and TP53−-AFPL group had not reached the median OS time.
Serial ctDNA monitoring and clinical efficacy of interventional therapy
In our enrolled patients, 12 patients had more than once interventional therapies and serial ctDNA were sampled at each therapy evaluation when available. Disease progression was observed in four patients, and seven patients were in stable disease and one patient was in partial remission (Figure 4A). In most patients (84%, 10/12), dynamic change of ctDNA abundance was consistent with the trends of tumor size except P009 and P039. ctDNA increased inconsistently when no evidence of AFP level and tumor size showed progress disease after TRCE treatment of P009 and P039 (http://fp.amegroups.cn/cms/a87146be21838a1704d8d51d2e52c465/JGO-20-409-8.xlsx). Notably, at next evaluating, a new lesion recurrent in liver was detected in P009 and P039 (Figure 4B).
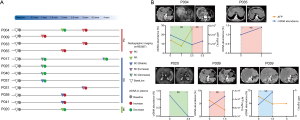
Blood TMB analysis of liver cancer
Recent studies had reported TMB as a predict factor in immunotherapy of pan-cancer (22), potentially in HCC (18). We further analyzed whether bTMB (blood TMB) could serve as an alternative biomarker of tTMB (tissue TMB), since biopsy tissue was unavailable when advanced liver cancer diagnosed. Median TMB was 6 and 5.76 mutations/Mb in tissue (28 evaluable samples) and blood (42 evaluable samples), respectively. To further analyze ctDNA could feasibility replace biopsy in calculated TMB, we analyzed 28 patients with paired tTMB and bTMB. The fraction of high TMB (TMB-H) was 17.8% (5/28) (cutoff =7.2 in geneplus database). bTMB could achieved 80% (4/5) positive predictive value and 91% (21/23) negative predictive value. bTMB showed a positive consistency with tTMB (Pearson r=0.7, P<0.0001) (Figure 5A). Moreover, patients with TP53 mutation had a higher TMB in both tissue (P=0.0084) and plasma samples (P=0.0038) (Figure 5B,C).

Discussion
Currently, the early diagnosis of liver cancer is still a worldwide problem. AASLD recommended diagnosis based on imaging techniques and/or biopsy (6). A previous meta-analysis reported that the sensitivity of ultrasonography for the detection of early-stage HCC was very limited in HCC-high risk group (23). Addition of AFP to ultrasonography increased the sensitivity of detection, but still suboptimal (23). Previous studies demonstrated cfDNA concentration in liver cancer patients were significantly higher than a health cohort, and ctDNA had a high sensitivity and specificity in earlier detection in non-metastatic liver cancer patients (9,10). ctDNA was non-invasive and could also overcome the temporal and spatial heterogeneity of tumor tissues. However, trace amounts of ctDNA in the early or mid-stage patients also brought challenges to the detection technology. Previous study showed the content of ctDNA in the plasma of liver cancer patient was about 5 ng/mL generally (24).
Our study showed a ctDNA positive rate of more than 95% (40/42) patients in pre-treatment plasma and obtained a consistency of 67.5% compared with matched tissue samples, which may be limited by tumor heterogeneity and subclonal mutational architecture. In this cohort, CV of ctDNA abundance is much less than that of AFP. ctDNA abundance presented more relevant with tumor load than AFP (Pearson r=0.7, AUC =0.82). These results suggested ctDNA was potential available and stable biomarker in auxiliary clinical diagnosis and evaluate tumor load of liver cancer.
Interventional therapy (TACE, RFA, TARE, I125 implantation, etc.) is one of standard therapy in advanced liver cancer patients. BCLC stage or PVTT status were evaluated as indicators for prognosis of interventional treatment (25). BCLC stage was related to ctDNA abundance. Further, better therapeutic effect of interventional therapy was observed in subgroup of negative detection of TP53, a cancer suppressor gene with the highest frequency mutate gene in liver cancer. In addition, considering drug perfusion may limit image method in accuracy of tumor size evaluating, whether ctDNA could be used in monitor treatment efficacy of liver cancer was clinically concerned, since it had been reported to be potential feasible in other solid tumors. In 84% patients, dynamic change of ctDNA abundance consistent with the mRECIST standard. For the other two patients, ctDNA was demonstrated to be more sensitive than image method, with a leading time of 45 days. To our knowledge, this is the first prospective study to reveal that targeted capture sequencing of ctDNA could be a potential evaluation factor in prognosis and clinical efficacy of interventional therapy.
In advanced HCC, there is an unsatisfied need for effective systemic therapies for advanced HCC, immunotherapy (IO) provides encouraging early indications of efficacy in advanced HCC (18). TMB of both tissue and blood had been demonstrated to be a useful biomarker for immunotherapy (26). In this study, tTMB and bTMB of advanced liver cancer patients were highly consistent. TP53 mutated patients had higher TMB levels, in both plasma and tissue samples. Our results led to the preliminary exploration of biomarker studies for immunotherapy in liver cancer, and showed ctDNA analysis as a better alternative method to assess TMB.
ctDNA abundance had been demonstrated to be related with tumor burden in other solid tumors, and as a biomarker in prognosis and progression with more sensitivity and specificity (16,27). TP53 mutation from circulating DNA also could be used to predicted prognosis. Although there was no significance in OS between the two group, the trend could be seen, and the median survival time of TP53 wildtype patients had not reach the median survival time. TP53 combination with AFP could distinguish the patients’ prognosis very well. Especially in baseline AFP <400 ng/mL patients, it is difficult to judge the prognosis of these cohorts, and after combining with TP53 mutation profiling, they can be distinguished well. Although our results were not statistically different, the differences survival trend could be seen between the two groups. We have reason to believe that the reason for the difference is because of our small sample size. Moreover, we will use a larger sample size to verify this conclusion. Although our study was limited by a small cohort size, However, clinical utility of ctDNA in the interventional treatment of advanced liver cancer will be confirmed.
In conclusion, evaluation of efficacy of interventional therapy for advanced liver cancer mainly based on AFP and imaging, which were challenged in accuracy. As a proof-of-concept study, we provide evidence that based on targeted capture sequencing, detecting ctDNA abundance may be a reliable method to assist in the diagnosis, prognosis and efficacy evaluation of advanced liver cancer. Considering that biopsy tissue was unnecessary when advanced liver cancer diagnosed, ctDNA may be an ideal biomarker for evaluating TMB before patients received immunotherapy.
Conclusions
ctDNA may be a reliable biomarker to assist in diagnosis and evaluation of prognosis and treatment efficacy in advanced liver cancer. Considering that biopsy is unnecessary when advanced liver cancer is diagnosed, ctDNA may be an ideal biomarker for evaluating tumor mutation burden prior to immunotherapy.
Acknowledgments
The authors wish to thank the patients and their families.
Funding:This work was supported by National Key Research and Development Program of China (No. 2017YFA0205200), National Natural Science Foundation of China (No. 81571785, 81801811, 81771957, 81901857), and Natural Science Foundation of Guangdong Province, China (No. 2018A030313074).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-409
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-409
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-409). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Patients with advanced liver cancer were obtained from the Zhuhai and Shenzhen people’s hospital, China. The study protocols were approved by the institutional review boards and all patients signed a written consent form prior to the study. The use of the clinical specimens for research purposes was approved by the Institutional Research Ethics Committee. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Valery PC, Laversanne M, Clark PJ, et al. Projections of primary liver cancer to 2030 in 30 countries worldwide. Hepatology 2018;67:600-11. [Crossref] [PubMed]
- Sirica AE, Gores GJ, Groopman JD, et al. Intrahepatic Cholangiocarcinoma: Continuing Challenges and Translational Advances. Hepatology 2019;69:1803-15. [Crossref] [PubMed]
- Siu EH, Chan AW, Chong CC, et al. Treatment of advanced hepatocellular carcinoma: immunotherapy from checkpoint blockade to potential of cellular treatment. Transl Gastroenterol Hepatol 2018;3:89. [Crossref] [PubMed]
- Zhou GH, Sun JH, Zhang YL, et al. Transcatheter embolization of hepatocellular carcinoma with epirubicin-loaded DC beads in Chinese patients. Transl Cancer Res 2019;8:279-89. [Crossref]
- McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer 2005;93:387-91. [Crossref] [PubMed]
- Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020-2. [Crossref] [PubMed]
- Qiu Y, He J, Chen X, et al. The diagnostic value of five serum tumor markers for patients with cholangiocarcinoma. Clin Chim Acta 2018;480:186-92. [Crossref] [PubMed]
- Danese E, Montagnana M, Lippi G. Circulating molecular biomarkers for screening or early diagnosis of colorectal cancer: which is ready for prime time? Ann Transl Med 2019;7:610. [Crossref] [PubMed]
- Ng CKY, Di Costanzo GG, Tosti N, et al. Genetic profiling using plasma-derived cell-free DNA in therapy-naïve hepatocellular carcinoma patients: a pilot study. Ann Oncol 2018;29:1286-91. [Crossref] [PubMed]
- Xu H, Zhu X, Xu Z, et al. Non-invasive Analysis of Genomic Copy Number Variation in Patients with Hepatocellular Carcinoma by Next Generation DNA Sequencing. J Cancer 2015;6:247-53. [Crossref] [PubMed]
- Liao W, Yang H, Xu H, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget 2016;7:40481-90. [Crossref] [PubMed]
- Sun Y, Meng R, Tang H, et al. Design of a liver cancer-specific selector for the analysis of circulating tumor DNA. Oncol Lett 2019;17:5369-76. [PubMed]
- Furtado LV, Segal JP. Circulating Tumor DNA Testing for Liver Cancer. Cell Mol Gastroenterol Hepatol 2015;1:458-9. [Crossref] [PubMed]
- Nong J, Gong Y, Guan Y, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun 2018;9:3114. [Crossref] [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415.
- Hu ZY, Xie N, Tian C, et al. Identifying Circulating Tumor DNA Mutation Profiles in Metastatic Breast Cancer Patients with Multiline Resistance. EBioMedicine 2018;32:111-8. [Crossref] [PubMed]
- Xu J, Zhang Y, Jia R, et al. Anti-PD-1 Antibody SHR-1210 combined with Apatinib for Advanced Hepatocellular Carcinoma, Gastric or Esophagogastric Junction Cancer: An Open-label, Dose Escalation and Expansion Study. Clin Cancer Res 2019;25:515-23. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41.e23. [Crossref] [PubMed]
- Minagawa M, Makuuchi M. Treatment of hepatocellular carcinoma accompanied by portal vein tumor thrombus. World J Gastroenterol 2006;12:7561-7. [Crossref] [PubMed]
- Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology 2016;150:835-53. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Tzartzeva K, Obi J, Rich NE, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology 2018;154:1706-18.e1. [Crossref] [PubMed]
- Xu RH, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155-61. [Crossref] [PubMed]
- Chan SL, Chong CC, Chan AW, et al. Management of hepatocellular carcinoma with portal vein tumor thrombosis: Review and update at 2016. World J Gastroenterol 2016;22:7289-300. [Crossref] [PubMed]
- Chan TA, Yarchoan M, Jaffee E, et al. Development of Tumor Mutation Burden as an Immunotherapy Biomarker: Utility for the Oncology Clinic. Ann Oncol 2019;30:44-56. [Crossref] [PubMed]
- Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med 2018;379:1754-65. [Crossref] [PubMed]


