
GSTM1 and GSTT1 copy number variants and the risk to Thai females of hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignancy found in the world and 85% of cases occur in men (1). Overall, an estimated 748,300 new cases of HCC and 695,900 deaths occurred in 2008 (2). In Thailand, HCC is the most common cancer found in males with an incidence rate of 33.9/100,000 and it is the third most common cancer found in females 12.9/100,000 population (3).
However, the diagnosis and prognosis of HCC are still poor. The 5-year overall survival (OS) rate is about 26–44% after resection because patients who are diagnosed with HCC are at an advanced stage (4). The HCC DNA marker is necessary for the diagnosis and prognosis of the disease. HCC risk factors include chronic hepatitis, viral infections, cirrhosis, alcohol, smoking, and consuming food contaminated with the following fungal toxins: Aspergillus flavus and Aspergillus parasiticus (5).
Glutathione S-transferases (GSTs) are a superfamily of enzymes that play an important role in the detoxification of the carcinogens glutathione S-transferase M1 (GSTM1) and glutathione S-transferase T1 (GSTT1) (6). The null genotypes of GSTM1 and GSTT1 might lead to a complete loss of enzyme activity. GSTM1 null genotypes have been associated with susceptibility to lung cancer, bladder cancer and colorectal cancer (7-9). The GSTM1 null genotype was detected in 60% of the population in western countries (10,11). While the GSTT1 null genotype was detected in only 20% of the population in western countries (12) and it was associated as a risk factor for susceptibility to colorectal cancer and endometrial cancer (13,14).
Real-time polymerase chain reaction (PCR) was used to investigate the DNA copy number variants (CNVs) of GSTM1 and GSTT1 in Thai HCC patients. The correlations between the frequencies of the GST genes’ CNVs in the control group and the patients were examined by Binary Logistic Regression and the clinico-pathological features of HCC were analyzed by Chi-square test. The CNVs for the GSTs and the survival status were determined by the Kaplan-Meier survival curve. A P value of ≤0.05 was set as the criteria for statistical significance. It is hoped that the results of this research will serve as fundamental knowledge on the effect of these genes’ CNVs on Thai HCC patients.
Methods
Sample collection and DNA isolation
Forty-nine HCC DNA samples were collected from the National Cancer Institute of Thailand. The DNA was extracted from formalin-fixed, paraffin-embedded tissues. Sixty-six healthy individuals with no prior history of cancer were selected as the healthy control group. The DNA from the control group was extracted from peripheral blood using the High Pure PCR Template Preparation Kit (Roche Diagnostics, Germany).
Detection of GSTM1 and GSTT1 CNVs
The GSTM1 and GSTT1 CNVs were determined by multiplex real-time PCR using the Luna® Universal qPCR Master mix kit (Bio-Rad Laboratories, USA) and the β-globin gene was used as a reference gene. The PCR primers for GSTM1 were 5'-GAA CTC CCT GAA AAG CTA AAG C-3' and 5'-5'-CTT GGG CTC AAA TAT ACG GTG G-3', the primers for GSTT1 were 5'-GCC ATC CTG CTC TAC CTG AC-3' and 5'-TGC CAG GTA CTC ATC CAC AC-3', and the primers for the β-globin gene were 5'-AAC TTC ATC CAC GTT CAC C-3' and 5'-GAA GAG CCA AGG ACA GGT AC-3'.
The master mix (10 µL) consisted of 1× Luna® Universal qPCR Master mix, 25 ng of DNA sample and 0.5 µM of primers. Real-time PCR was performed by the BioRad CFX connect qPCR (real-time PCR) system using the following steps: initial denaturation at 95 °C for 1 minute, 40 cycles under denaturation conditions at 95 °C for 5 seconds, and primer annealing and polymerization at 60 °C for 30 seconds. The gene copy numbers were determined by the ∆Ct method = Ct, target gene – Ct, reference gene (15).
Statistical analysis
The frequencies of the CNVs in the control group and the patients were compared using the Chi-square test. Binary logistic regression was used to evaluate the relationship between the control and disease genotypes. The relationships between the patients’ clinico-pathological parameters (gender, stage, size of the tumor, differentiation, and age at diagnosis) to the GSTM1 and GSTT1 CNVs were examined by Chi-square test. Survival status was determined by the Kaplan-Meier survival method and the log-rank test. The patients were followed up for a period of 1–155 months. A P value of ≤0.05 was considered statistically significant.
Results
Detection of GSTM1 and GSTT1 CNVs
The GSTM1 CNVs were determined using the real-time PCR technique to analyze 49 DNA samples obtained from HCC patients and 66 DNA samples obtained from the control group. The fluorescent data in Figure 1 represents the Ct value and the melting curve. The results show that the GSTM1 for genotypes was 0/0 69.3% and 1/0 22.6%, 1/1 8.1% of cases. The GSTT1 for genotypes was 0/0 24.5% and 1/0 42.8%, 1/1 10.2%, >1 22.5% of cases.
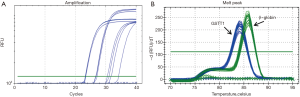
The relationship between the frequencies of GSTM1 and GSTT1 CNVs in the control group and patients
The GSTM1 and GSTT1 CNVs were analyzed. The results show that the difference between the controls and the patients was not statistically significant (P≥0.05). Overall, the frequencies of the CNVs for the GSTM1 genotypes 0/0 between patients and controls were 60.6% and 69.6%, respectively and the GSTT1 genotypes were 24.4% and 37.8%, respectively. Only the GSTT1 genotypes 0/0 were associated with an increased risk of hepatocellular carcinogenesis (OR value was 1.88). While the GSTM1 genotypes were not associated with the risk factors of HCC (OR value was 0.67). Table 1 shows a summary of the data.
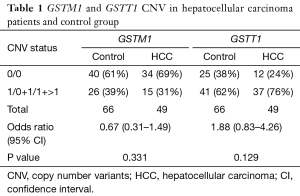
Full table
Statistical analysis of GSTM1 and GSTT1 CNVs and patients’ clinico-pathological parameters
The correlation between the patients’ clinico-pathological parameters and the CNVs of GSTM1 and GSTT1 were analyzed, as shown in Tables 2,3. The GSTM1 CNVs were associated with the gender of the patients (P=0.002). Stage, the size of the tumor, differentiation, and the patient’s age at diagnosis showed no significant difference (P>0.05). However, in the case of the GSTT1 CNVs, no correlations were found to any of the clinico-pathological parameters (P>0.05).
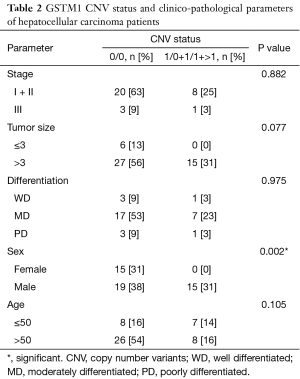
Full table
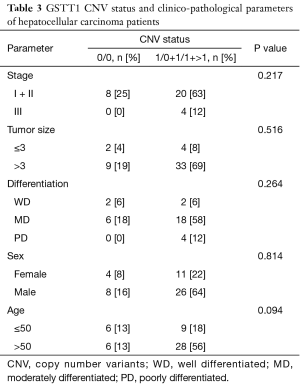
Full table
Survival of patients with GSTM1 and GSTT1 CNVs
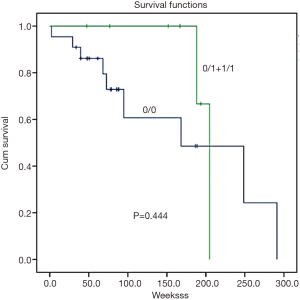
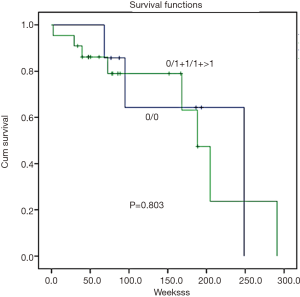
Survival analysis was determined by the Kaplan-Meier survival curve. In summary, there was no correlation found between GSTM1 and GSTT1 CNVs and patient survival (Figures 2,3, P=0.444 and 0.803, respectively).
Discussion
GSTM1 and GSTT1 genes are involved in the detoxification of activated carcinogen and other toxins. When this happens, the loss of enzyme catalytic activity from the deletion genotype leads to a decrease of cell protection from various toxic agents (16) causing an association with susceptibility to many cancers (7-9).
While, GSTM1 has been associated with an increased risk of lung (7) and colorectal cancer (17) in previous research. We found no significant difference between controls and patients for the GSTM1 CNVs in HCC; however, we found that GSTT1 genotypes 0/0 increased the risk of susceptibility to HCC (OR =1.88). This is in parallel with other research on lung, breast, colon and endometrial cancer, which concluded that GSTT1 genotypes 0/0 or null genotypes resulted in a loss of GSTT1 enzyme activity and reduced toxin detoxification leading to an increased risk of susceptibility to carcinogenesis (13,14,18,19).
First, the gene copy numbers for GSTM1 and GSTT1 were determined from 49 HCC samples by the real-time PCR technique. Then, the associations with clinico-pathological parameters were analyzed. The results show that only the GSTM1 CNVs were associated with the gender of patients (P=0.002), the frequency of genotypes 0/0 was more dominant in females than males (male 55.9%, female 100.0%). The results of this research concur with a study on Japanese lung cancer patients (20).
This phenomenon allows sex hormones to play a role in protecting organs such as the lung and liver from inflammatory substances and preventing HCC development in females (21). The protective effect of estrogen inhibited proinflammatory cytokines such as IL-6 contributed to the causes of liver disease (22) and the risk of female HCC incidence increased at the menopausal age (23). In addition, inflammation resulting from oxidative substances may be prevented by the GSTM1 function; however, the data assumed that the null genotype of the GSTM1 gene would increase the risk of inflammatory diseases (24). Prior research, which supports our results, identified GSTM1 CNVs and sex hormones as two factors that were associated with liver inflammation and carcinogenesis. GST polymorphisms were found to influence the efficiency of estrogen metabolism in women (25).
According to previous research, GSTM1 CNVs have been associated with the progression of phenotypes in colorectal cancer (26) and oral squamous cell carcinoma (27). GSTM1 CNVs tended to be related to tumor size (P=0.077). However, we found no correlation between the GSTT1 CNVs and clinico-pathological status.
In conclusion, the results of this research indicate that only the GSTT1 CNVs are associated with an increased risk of susceptibility to HCC. GSTM1 copy numbers were also associated with the gender of patients more dominantly in females than in males and tended to be related to the progression phenotype. No association was found between the GSTT1 CNVs and the clinico-pathological status of patients.
Acknowledgements
The authors would like to thank the Rangsit University Research Fund, the Faculty of Science, Rangsit University and National Cancer Institute, Thailand.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Rangsit University Ethics Committee (No. RSPE 20/2560) and Ethics Committee of National Cancer Institute, Thailand (No. EC COA 005/2017). As this was a retrospective study, informed consent is not required.
References
- Song J, Wang LZ, Li X, et al. Polymorphisms of vascular endothelial growth factor on prognosis in hepatocellular carcinoma patients receiving transcatheter arterial chemoembolization treatment. Genet Mol Res 2014;13:8946-53. [Crossref] [PubMed]
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- National Cancer Institute: Hospital based cancer registry annual report 2013. BTS PRESS Co., LTD, Bangkok, 2015.
- Wu KT, Wang CC, Lu LG, et al. Hepatocellular carcinoma: clinical study of long-term survival and choice of treatment modalities. World J Gastroenterol 2013;19:3649-57. [Crossref] [PubMed]
- Santos NP, Colaço AA, Oliveira PA. Animal models as a tool in hepatocellular carcinoma research: A Review. Tumour Biol 2017;39:1010428317695923. [Crossref] [PubMed]
- Boyer TD. The glutathione S-transferases: an update. Hepatology 1989;9:486-96. [Crossref] [PubMed]
- Hirvonen A, Husgafvel-Pursiainen K, Anttila S, et al. The GSTM1 null genotype as a potential risk modifier for squamous cell carcinoma of lung. Carcinogenesis 1993;14:1479-81. [Crossref] [PubMed]
- Nørskov MS, Frikke-Schmidt R, Bojesen SE, et al. Copy number variantsin glutathione-S-transferaseT1 and M1 predicts incidence and 5-year survival from prostate and bladder cancer, and incidence of corpus uteri cancer in the general population. Pharmacogenomics J 2011;11:292-9. [Crossref] [PubMed]
- Loktionov A, Watson MA, Gunter M, et al. Glutathione S-transferase gene polymorphism in colorectal cancer patients: interaction between GSTM1 and GSTM3 allele variants as a risk-modulating factor. Carcinogenesis 2001;22:1053-60. [Crossref] [PubMed]
- Xu S, Wang Y, Roe B, et al. Characterization of the human class Mu Glutathione S-transferase gene cluster and GSTM1 deletion. J Biol Chem 1998;273:3517-27. [Crossref] [PubMed]
- Pearson WR, Vorachek WR, Xu SJ, et al. Identification of class Mu Glutathione S-transferase genes GSTM1- GSTM5 on human chromosome 1p13. Am J Hum Genet 1993;53:220-33. [PubMed]
- Naoe T, Tagawa Y, Kiyoi H, et al. Prognostic significance of the null genotype of glutathione S-transferase-T1 in patients with acute myeloid leukemia: increased early death after chemotherapy. Leukemia 2002;16:203-8. [Crossref] [PubMed]
- Chenevix-Trench G, Young J, Coggan M, et al. Glutathione S-transferase M1 and T1 polymorphism: susceptibility to colon cancer and age of onset. Carcinogenesis 1995;16:1655-7. [Crossref] [PubMed]
- Karageorgi S, Prescott J, Wong JY, et al. GSTM1 and GSTT1 copy number variation population-based studies of endometrial cancer risk. Cancer Epidemiol Biomarkers Prev 2011;20:1447-52. [Crossref] [PubMed]
- Covault J, Abreu C, Kranzler H, et al. Quantitative real-time PCR for gene dosage determinations in microdeletion genotypes. Biotechniques 2003;35:594-6, 598. [Crossref] [PubMed]
- Norppa H. Cytogenetic biomarkers and genetic polymorphisms. Toxicol Lett 2004;149:309-34. [Crossref] [PubMed]
- Wang J, Jiang J, Zhao Y, et al. Genetic polymorphisms of glutathione S-transferase genes and susceptibility to colorectal cancer: a case-control study in an Indian population. Cancer Epidemiol 2011;35:66-72. [Crossref] [PubMed]
- El-Zein R, Abdel-Rahman SZ, Sankar A, et al. Genetic polymorphism and risk for development of lung cancer. Environ Mol Mutagen 1996;27:20.
- Kimi L, Ghatak S, Yadav RP, et al. Relevance of GSTM1, GSTT1 and GSTP1 Gene Polymorphism to Breast Cancer Susceptibility in Mizoram Population, Northeast India. Biochem Genet 2016;54:41-9. [Crossref] [PubMed]
- Kihara M, Noda K, Kihara M. Distribution of GSTM1 null genotype in relation to gender, age and smoking status in Japanese lung cancer patients. Pharmacogenetics 1995;5:S74-9. [Crossref] [PubMed]
- El Mahdy Korah T, Abd Elfatah Badr E, Mohamed Emara M, et al. Relation between sex hormones and hepatocellular carcinoma. Andrologia 2016;48:948-55. [Crossref] [PubMed]
- Shi L, Feng Y, Lin H, et al. Role of estrogen in hepatocellular carcinoma: is inflammation the key? J Transl Med 2014;12:93. [Crossref] [PubMed]
- Liu P, Xie SH, Hu S, et al. Age-specific sex difference in the incidence of hepatocellular carcinoma in the United States. Oncotarget 2017;8:68131-7. [PubMed]
- Wu W, Peden D, Diaz-Sanchez D. Role of GSTM1 in resistance to lung inflammation. Free Radic Biol Med 2012;53:721-9. [Crossref] [PubMed]
- Henningson MC, Hietala M, Bågeman E, et al. Interactions between oral contraceptive status and GSTM1 and GSTT1 deletions on insulin-like growth factor-1 (IGF-1) plasma levels in young healthy women. Growth Horm IGF Res 2010;20:432-7. [Crossref] [PubMed]
- Pongtheerat T, Chaksangchaichot P, Saelee P. Prognosis in Thai colorectal-cancer patients with GSTM1 and GSTT1 copy number variation. Eur J Oncol 2013;18:189-95.
- Liu CJ, Chang CS, Lui MT, et al. Association of GST genotypes with age of onset and lymph node metastasis in oral squamous cell carcinoma. J Oral Pathol Med 2005;34:473-7. [Crossref] [PubMed]


