Molecular profiling of biliary tract cancer: a target rich disease
Introduction
Biliary tract cancer (BTC) classification
BTCs consist of tumors that arise from the epithelial cells lining the biliary tree and can be either intrahepatic cholangiocarcinoma (IHCCA), extrahepatic cholangiocarcinoma (EHCCA) or gallbladder cancer (GBC). EHCCAs are further classified as Klatskin (hilar) and distal cholangiocarcinomas (1). BTCs are heterogeneous cancers that are increasing in incidence worldwide, are often refractory to standard chemotherapy regimens and bear a poor prognosis. These cancers are associated with gallstones, inflammatory bowel disease, obesity, hepatitis, congenital cystic lesions in the bile duct or toxin exposure. The genetic alterations in BTC result in the activation of complex molecular downstream signaling pathways, altered chromatin remodeling, deregulation of DNA repair and accelerated angiogenesis (2). BTCs are rich in actionable genetic aberrations (GA) and it is possible now to identify unique molecular subsets of BTC that can be effectively treated with an individualized approach.
Genomic landscape based on tumor location (IHCCA, EHCCA and GBC)
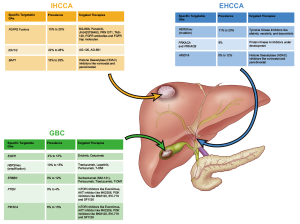
There has been a rising interest in the genomic landscape of BTCs with the advent of new, advanced technologies like next-generation sequencing (NGS). It has been observed that the mutational profile of BTCs vary with location of the tumor. Isocitrate dehydrogenase 1 or 2 (IDH1/2), BAP1 mutations and FGFR2 fusions are more commonly observed in IHCCA, on the other hand in EHCCA, KRAS, p53 and SMAD4 mutations are more frequent. Activating ERBB2 and ERBB3 mutations and inactivating PTEN and TSCI mutations are seen in GBC (1,3,4). Common actionable GA, their prevalence and possible targeted treatment options based on the location of the tumor are depicted in Figure 1.
Genomic landscape differences seen in different patient populations (West versus East)
The etiology of the BTCs varies in different parts of the world. GBC in Asia and Latin America is associated with cholelithiasis whereas in Western Europe and North America, GBC is becoming very uncommon with the increasing use of laparoscopic cholecystectomy for symptomatic gallstones (5). In the case of cholangiocarcinoma in Southeast Asia, parasitic infection by the liver fluke, Opisthorchis viverrini and hepatitis B are often responsible (6,7). On the other hand in the western world, obesity, inflammatory bowel disease with sclerosing cholangitis, hepatitis C and toxin exposure have a pathogenic role (7). It follows that there is also considerable genetic diversity between these cancers corresponding to the geographic region of occurrence.
Ong et al. performed whole exome sequencing of liver-fluke associated cholangiocarcinoma from Asian patients and reported mutations in P53, KRAS, MLL3, ROBO2, RNF43, PEG3 and GNAS genes. The latter groups of genes are involved in the deactivation of histone modifiers, activation of G protein signaling and loss of genome stability (8). The same group then sequenced non-liver fluke associated cholangiocarcinoma and noted that there was a significantly higher prevalence of P53, SMAD4, MLL3 and GNAS in liver fluke associated cholangiocarcinoma while non-liver fluke associated cholangiocarcinomas had a higher rate of IDH1/2 and BAP1 mutations (8). Similar findings were reported from an exome sequencing study from western patients with cholangiocarcinoma with a higher frequency of mutations in the chromatin modulating genes, including ARID1A, BAP1 and PBRM1 being reported (9). Mutations in the FGFR family, including FGFR gene fusions/translocations have been reported in up to 10% of IHCCAs in the western patients while these mutations have not been reported to our knowledge in the liver fluke associated cholangiocarcinomas (3,10). It is evident, based on the above observations that genetic differences exist depending upon the etio-pathogenesis of cholangiocarcinoma. However, tumor location may at least partly account for these differences. Liver fluke associated cholangiocarcinoma for instance, is more likely to be extrahepatic (distal or perihilar) in location (11), which as discussed has a different genetic profile as compared with IHCCA.
Frequently dysregulated pathways
HER2/neu, HER1/EGFR signaling pathways
HER2/neu gene is a key driver of tumorigenesis in several solid tumors, including breast, non-small cell lung cancer and colorectal cancer and its overexpression as a result of gene amplification indicates an adverse prognosis. Our group recently studied HER2/neu protein expression in 187 cases of GBC using the accepted College of American Pathologists (CAP)/American Society of Clinical Oncology (ASCO) criteria and noted a 13% HER2/neu overexpression rate 3+ by immunohistochemistry (4). Yoshikawa and colleagues reported a 0.9% and 8.5% incidence of HER2/neu protein overexpression in intra- and EHCCA, respectively. In their series, early stage disease and well-differentiated tumors had a higher incidence of HER2/neu positivity (12). Data with regards to the prognostic value of HER2/neu overexpression in biliary cancers is mixed, with some studies suggesting a worse prognosis (13,14), while others suggest a better prognosis (12,15). Although the data regarding HER2/neu targeted therapy with trastuzumab in biliary cancer is limited, smaller case series indicate therapeutic benefit in GBC patients who had HER2/neu amplification but larger studies should be conducted to validate these findings (16). HER2/neu mutations in the kinase domain occur more commonly in EHCCA and maybe subjects for irreversible tyrosine kinase inhibitors (TKIs) targeting HER2/3 and EGFR, such as afatinib, neratinib, and dacomitinib (3).
Abnormal activation of EGFR receptors, leads to activation of downstream mitogen-activated protein kinase (MAPK)/ERK and phosphatidylinositol 3-kinase (PI3K)/PTEN/AKT pathways, both of which are well-established oncogenic pathways in BTC (17). A phase II study of erlotinib in advanced BTC indicated that EGFR expression was noted in 81% of cases of whom 17% experienced disease stability for 6 months (18). Lubner et al. further studied this signal in a multicenter phase II study of erlotinib and bevacizumab; of the 53 patients enrolled; 12% experienced partial response and stability was seen in 51% (19). A subsequent phase III study of gemcitabine, oxaliplatin ± erlotinib, showed no difference in overall survival between these two groups, however there was an improved progression-free survival and response rate in the erlotinib arm (20). These studies indicate that EGFR targeting in BTC continues to be an area of clinical value.
BRAF mutations occur in 5% of BTC cases, particularly in IHCCA (21-24). Impressive activity was noted in metastatic melanoma with BRAF V600 mutations with a combination of dabrafenib (BRAF inhibitor) and trametinib (MEK1 and MEK2 inhibitor) (25). Loaiza-Bonilla et al. reported an impactful response in an IHCCA patient who received this dual therapy, using a combination of BRAF inhibition and a MEK inhibitor (26). This indicates the need for prospective clinical trials for BTCs patients with mutant BRAF.
FGFR signaling pathway
FGFR pathway GAs, particularly FGFR fusion genes or translocations have been described in 10–16% of IHCCA and have gained a lot of attention as potential targets for therapeutic intervention (3,27). FGFR fusions are very uncommon in extrahepatic or GBC and are almost exclusively seen in IHCCA only. The FGFR family consists of four transmembrane receptors (FGFR 1–4), 22 FGFR ligands and a heparan sulfate proteoglycan (HSPG) that stabilizes and sequesters the FGFs. The major downstream signaling routes for FGFR are through the Ras-Raf-MAPK, PI3K pathways and the protein-serine/threonine kinase AKT. Dysregulation of the FGFR pathway can occur through FGFR mutations, translocations, amplification, overexpression and alteration of regulatory influences such as by FGF ligand amplification. FGFR amplifications are rare in BTC but FGFR2 mutations and FGFR2 fusions occur relatively frequently in IHCCA (27). Sia et al. reported that FGFR2-PPHLN1 fusions occur in up to 16% of IHCCA (28). In our experience, FGFR2-BICC fusion was the commonest fusion noted. Our group has investigated the prognostic role of FGFR pathway GAs. FGFR2 GA, including fusion genes appears to be associated with a relatively indolent clinical disease course of cholangiocarcinoma (29). The advent of FGFR-targeted therapies has incentivized further clinical research in this field. These therapies include multi-TKIs that also inhibit FGFR (such as ponatinib, nintedanib, dovitinib and brivanib), specific FGFR directed small molecule TKI (BGJ398, JNJ425756493, PRN1371, ARQ087, TAS-120) FGFR antibodies and FGFR trap molecules. Phase II BGJ398 results were reported recently; 50 patients with BTC having FGFR genetic alterations were enrolled, the majority being IHCCA. The overall response rate was 15%, disease control rate was 95% with progression-free survival of 6 months, indicating that FGFR-directed therapy is a successful investigational strategy for FGFR mutated cholangiocarcinoma (30).
IDH1/2 mutations in BTC
In 2008, mutations in IDH1 and IDH2 were first identified in glioma and acute myeloid leukemia. Subsequently, these were identified in other solid tumors including cholangiocarcinoma (31). In IHCCA, an estimated 20% have IDH1 while 5% have IDH2 mutations (32). These mutations are not seen in EHCCA or GBC (3,33). The most common IDH mutations occur in arginine residues in the catalytic pockets: IDH1 (R132) and IDH2 (R172 or R140) (34). IDH1 and IDH2 catalyzes the NADP+-dependent reversible conversion of isocitrate to α-ketoglutarate, whereas gain-of-function IDH1 or 2 mutations catalyze the oxidation of α-ketoglutarate to 2 dihydroxyglutarate (2-HG), using NADPH as a cofactor (35). It is believed that 2-HG is mutagenic although the mechanism is not well understood. However, this alteration causes change in redox potential, increased CpG island and histone methylation, thus blocking cell differentiation, impaired collagen maturation and basement membrane formation. Mutant IDH promotes carcinogenesis by blocking hepatocyte differentiation and increasing pools of hepatic progenitors that are susceptible to additional oncogenic hits (36). IDH1 mutation is regarded as a favorable prognostic factor in glioma. However, we and others have not found any prognostic significance to IDH1/2 mutation in cholangiocarcinoma (32,37). Small molecule specific inhibitors against IDH1/2 may have promising anti-tumor activity in this subgroup. Recently Burris et al. reported the findings of a dose-escalation study of AG-120 in various cancer types having these mutations. Of the 20 cholangiocarcinoma patients enrolled, response or stability was noted in 12 patients with the disease stability demonstrable beyond 6 months (38). Other allosteric IDH inhibitors like AG-881, are currently under development for these cancers (39).
DNA repair mutations in BTC
DNA repair mechanisms are essential for maintaining genomic stability and defects in these occur in BTC. Gene mutations leading to defective DNA mismatch repair (MMR) are commonly seen in several solid tumors like colorectal cancer, endometrial and gastric cancer (40,41). Data regarding presence of MMR in BTC is limited. Goyal et al. reported a 9% rate of MMR protein loss in cholangiocarcinoma patients on immunohistochemistry, with 4.5% patients being microsatellite instability (MSI)-high (42). In our series of 321 BTC who underwent mutational profiling, DNA repair mutations (MSH6, BRCA1, BRCA2, ATM, MLH1 or MSH2 genes) occurred in 13% IHCCA, 26% in EHCCA and 6% of GBC cases (32). The subset of cancers with MMR system defects is very sensitive to programmed cell death protein 1 (PD-1) blockade using checkpoint inhibitor agents like pembrolizumab (43). Specific DNA repair inhibitors targeting homologous recombination repair, base excision repair and nucleotide excision repair are under investigation currently. BTC patients with mutations in the DNA repair pathways can represent a subset wherein targeted therapeutics in the form of specific DNA repair inhibitors or immunotherapy may be effective. Further clinical trials are needed to validate the safety and efficacy of these treatment modalities.
Mutations in chromatin remodeling genes
Inactivating genetic alterations in ARID, BAP1, PBRM and MLL that are responsible for chromatin remodeling have been noted in renal cell carcinoma uveal melanoma, ovarian, colorectal and gastric cancers. Chromatin remodeling allows genomic DNA to access regulatory transcriptional proteins and thereby controls gene expression. GA in this process have recently been implicated in the development of BTC (9). BAP1 encodes for a nuclear deubiquitinase, while PBRM1 and ARID1A both encode a subunit of the ATP dependent SWI/SNF chromatin-remodeling complexes (44,45). Jiao et al. observed that mutations in at least one of these genes occurred in almost half of the BTCs sequenced in their study (9). The prognostic role of mutations in chromatin remodeling genes is currently unknown, though BAP1 mutations were associated with aggressive disease resulting in bony metastases (3). Another case series reported 22 cases of cholangiocarcinoma with BAP1 mutation and noted that 13 (59%) had bone metastases at presentation. These patients experienced rapid tumor progression with a mean time for tumor progression of 3.8 months (46). This study highlights the dire need to focus on BAP1-directed targeted therapies as currently there are no effective therapies for cholangiocarcinoma with these mutations. It is hypothesized that histone deacetylase (HDAC) inhibitors such as vorinostat and panobinostat may offer therapeutic value and need to be explored further prospectively (47).
Molecular classification with prognostic staging system/score for BTC
Cancer classification has traditionally relied upon pathologic criteria that are based on site of origin. Recent sequencing studies have highlighted that disparate cancers arising from different organ systems may harbor similar GA causing them to exhibit a clinical course and targeted therapy response that is compatible with the molecular phenotype rather than the organ of origin. For instance BRAF mutant cholangiocarcinoma may have clinical homology with melanoma having the BRAF mutation rather than with FGFR mutant cholangiocarcinoma. Larger genomic studies conducted with The Cancer Genome Atlas (TCGA) research in lung, colorectal, pancreas, breast, bladder, endometrial and other cancers have revealed that each of these cancers can be further subdivided into three or four more molecular subtypes based on recurrent genetic alterations that are commonly expressed. In gastric cancer, four molecular subgroups have been identified: (I) Epstein-Barr virus (EBV) type (high PIK3CA mutation, hypermethylation, expression of PDL1; (II) MSI subtype; (III) genomically stable subtype (diffuse histology, RAS mutation and genes encoding adhesion proteins); and (IV) tumors with chromosomal instability (intestinal type histology, aneuploidy and receptor TKI amplification) (48). In case of pancreatic cancer, similarly four subgroups have been proposed: (I) stable (20% aneuploidy); (II) locally rearranged (30%, focal event in one or two chromosomes); (III) scattered (36%, <200 structural variation events); and (IV) unstable (14%, >200 structural variation events, defects in DNA repair and maintenance) (49). This classification has important implications for targeted and immunotherapy. For instance, the MSI subtype and the EBV type of gastric cancers are sensitive to immune inhibition with checkpoint blockers. The ‘unstable’ variant of pancreatic cancer may respond well to DNA repair blockers, such as inhibitors of poly(ADP-ribose) polymerase (PARP). TCGA analysis of cholangiocarcinoma is now complete and the expectation is that we will have a similar molecular subtyping of this disease that may result in prognostic and therapeutic stratification.
In summary, molecular analysis of BTC has been extremely instructive; this disease has considerable genetic heterogeneity that can be effectively targeted with novel agents. The plethora of actionable mutations in these orphan diseases has made precision medicine a reality.
Acknowledgements
This study was supported by Paula and Jeff Laut Foundation, Graeme and Lee Dayton Foundation. Illustrations by David Aten, MA, senior medical illustrator MD Anderson.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003-10. [Crossref] [PubMed]
- Sia D, Tovar V, Moeini A, et al. Intrahepatic cholangiocarcinoma: pathogenesis and rationale for molecular therapies. Oncogene 2013;32:4861-70. [Crossref] [PubMed]
- Churi CR, Shroff R, Wang Y, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 2014;9:e115383. [Crossref] [PubMed]
- Roa I, de Toro G, Schalper K, et al. Overexpression of the HER2/neu Gene: A New Therapeutic Possibility for Patients With Advanced Gallbladder Cancer. Gastrointest Cancer Res 2014;7:42-8. [PubMed]
- Stinton LM, Shaffer EA. Epidemiology of gallbladder disease: cholelithiasis and cancer. Gut Liver 2012;6:172-87. [Crossref] [PubMed]
- Rizvi S, Borad MJ, Patel T, et al. Cholangiocarcinoma: molecular pathways and therapeutic opportunities. Semin Liver Dis 2014;34:456-64. [Crossref] [PubMed]
- Tyson GL, El-Serag HB. Risk factors for cholangiocarcinoma. Hepatology 2011;54:173-84. [Crossref] [PubMed]
- Ong CK, Subimerb C, Pairojkul C, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet 2012;44:690-3. [Crossref] [PubMed]
- Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 2013;45:1470-3. [Crossref] [PubMed]
- Graham RP, Barr Fritcher EG, Pestova E, et al. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol 2014;45:1630-8. [Crossref] [PubMed]
- Wiwanitkit V. Clinical findings among 62 Thais with cholangiocarcinoma. Trop Med Int Health 2003;8:228-30. [Crossref] [PubMed]
- Yoshikawa D, Ojima H, Iwasaki M, et al. Clinicopathological and prognostic significance of EGFR, VEGF, and HER2 expression in cholangiocarcinoma. Br J Cancer 2008;98:418-25. [Crossref] [PubMed]
- Settakorn J, Kaewpila N, Burns GF, et al. FAT, E-cadherin, beta catenin, HER 2/neu, Ki67 immuno-expression, and histological grade in intrahepatic cholangiocarcinoma. J Clin Pathol 2005;58:1249-54. [Crossref] [PubMed]
- Nakazawa K, Dobashi Y, Suzuki S, et al. Amplification and overexpression of c-erbB-2, epidermal growth factor receptor, and c-met in biliary tract cancers. J Pathol 2005;206:356-65. [Crossref] [PubMed]
- Shafizadeh N, Grenert JP, Sahai V, et al. Epidermal growth factor receptor and HER-2/neu status by immunohistochemistry and fluorescence in situ hybridization in adenocarcinomas of the biliary tree and gallbladder. Hum Pathol 2010;41:485-92. [Crossref] [PubMed]
- Javle M, Churi C, Kang HC, et al. HER2/neu-directed therapy for biliary tract cancer. J Hematol Oncol 2015;8:58. [Crossref] [PubMed]
- Andersen JB, Thorgeirsson SS. Genetic profiling of intrahepatic cholangiocarcinoma. Curr Opin Gastroenterol 2012;28:266-72. [Crossref] [PubMed]
- Philip PA, Mahoney MR, Allmer C, et al. Phase II study of erlotinib in patients with advanced biliary cancer. J Clin Oncol 2006;24:3069-74. [Crossref] [PubMed]
- Lubner SJ, Mahoney MR, Kolesar JL, et al. Report of a multicenter phase II trial testing a combination of biweekly bevacizumab and daily erlotinib in patients with unresectable biliary cancer: a phase II Consortium study. J Clin Oncol 2010;28:3491-7. [Crossref] [PubMed]
- Lee J, Park SH, Chang HM, et al. Gemcitabine and oxaliplatin with or without erlotinib in advanced biliary-tract cancer: a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2012;13:181-8. [Crossref] [PubMed]
- Simbolo M, Fassan M, Ruzzenente A, et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget 2014;5:2839-52. [Crossref] [PubMed]
- Saetta AA, Papanastasiou P, Michalopoulos NV, et al. Mutational analysis of BRAF in gallbladder carcinomas in association with K-ras and p53 mutations and microsatellite instability. Virchows Arch 2004;445:179-82. [Crossref] [PubMed]
- Andersen JB, Spee B, Blechacz BR, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 2012;142:1021-31.e15. [Crossref] [PubMed]
- Tannapfel A, Sommerer F, Benicke M, et al. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut 2003;52:706-12. [Crossref] [PubMed]
- Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367:1694-703. [Crossref] [PubMed]
- Loaiza-Bonilla A, Clayton E, Furth E, et al. Dramatic response to dabrafenib and trametinib combination in a BRAF V600E-mutated cholangiocarcinoma: implementation of a molecular tumour board and next-generation sequencing for personalized medicine. Ecancermedicalscience 2014;8:479. [Crossref] [PubMed]
- Hierro C, Rodon J, Tabernero J. Fibroblast Growth Factor (FGF) Receptor/FGF Inhibitors: Novel Targets and Strategies for Optimization of Response of Solid Tumors. Semin Oncol 2015;42:801-19. [Crossref] [PubMed]
- Sia D, Losic B, Moeini A, et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat Commun 2015;6:6087. [Crossref] [PubMed]
- Jain A, Shroff RT, Kelley RK, et al. FGFR pathway genetic aberrations in cholangiocarcinoma: Demographics and experience with targeted therapy. J Clin Oncol 2016;34:abstr 109.
- Javle MM, Shroff RT, Zhu A, et al. A phase 2 study of BGJ398 in patients (pts) with advanced or metastatic FGFR-altered cholangiocarcinoma (CCA) who failed or are intolerant to platinum-based chemotherapy. J Clin Oncol 2016;34:abstr 335.
- Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov 2013;3:730-41. [Crossref] [PubMed]
- Ross JS, Wang K, Javle MM, et al. Comprehensive genomic profiling of biliary tract cancers to reveal tumor-specific differences and frequency of clinically relevant genomic alterations. J Clin Oncol 2015;33:abstr 4009.
- Borger DR, Goyal L, Yau T, et al. Circulating oncometabolite 2-hydroxyglutarate is a potential surrogate biomarker in patients with isocitrate dehydrogenase-mutant intrahepatic cholangiocarcinoma. Clin Cancer Res 2014;20:1884-90. [Crossref] [PubMed]
- Grassian AR, Pagliarini R, Chiang DY. Mutations of isocitrate dehydrogenase 1 and 2 in intrahepatic cholangiocarcinoma. Curr Opin Gastroenterol 2014;30:295-302. [Crossref] [PubMed]
- McKenney AS, Levine RL. Isocitrate dehydrogenase mutations in leukemia. J Clin Invest 2013;123:3672-7. [Crossref] [PubMed]
- Saha SK, Parachoniak CA, Bardeesy N. IDH mutations in liver cell plasticity and biliary cancer. Cell Cycle 2014;13:3176-82. [Crossref] [PubMed]
- Goyal L, Govindan A, Sheth RA, et al. Prognosis and Clinicopathologic Features of Patients With Advanced Stage Isocitrate Dehydrogenase (IDH) Mutant and IDH Wild-Type Intrahepatic Cholangiocarcinoma. Oncologist 2015;20:1019-27. [Crossref] [PubMed]
- Burris H, Mellinghoff I, Maher E, et al. The first reported results of AG-120, a first-in-class, potent inhibitor of the IDH1 mutant protein, in a Phase I study of patients with advanced IDH1-mutant solid tumors, including gliomas. Mol Cancer Ther 2015;14:abstract PL04-05.
- Merla A, Liu KG, Rajdev L. Targeted Therapy in Biliary Tract Cancers. Curr Treat Options Oncol 2015;16:48. [Crossref] [PubMed]
- Kim H, Jen J, Vogelstein B, et al. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol 1994;145:148-56. [PubMed]
- Oliveira C, Seruca R, Seixas M, et al. The clinicopathological features of gastric carcinomas with microsatellite instability may be mediated by mutations of different "target genes": a study of the TGFbeta RII, IGFII R, and BAX genes. Am J Pathol 1998;153:1211-9. [Crossref] [PubMed]
- Goyal L, Deshpande V, Chung DC, et al. Mismatch repair protein loss and microsatellite instability in cholangiocarcinoma. J Clin Oncol 2014;32:abstr 237.
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Wu RC, Wang TL, Shih IeM. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther 2014;15:655-64. [Crossref] [PubMed]
- Chiang NJ, Shan YS, Hung WC, et al. Epigenetic regulation in the carcinogenesis of cholangiocarcinoma. Int J Biochem Cell Biol 2015;67:110-4. [Crossref] [PubMed]
- Al-Shamsi HO, Anand D, Shroff RT, et al. BRCA-associated protein 1 mutant cholangiocarcinoma: an aggressive disease subtype. J Gastrointest Oncol 2016;7:556-61. [Crossref] [PubMed]
- Kwak TW. Antitumor activity of vorinostat-incorporated nanoparticles against human cholangiocarcinoma cells. J Nanobiotechnology 2015;13:60. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]


