The role of EGFR monoclonal antibodies (MoABs) cetuximab/panitumab, and BRAF inhibitors in BRAF mutated colorectal cancer
Background
Metastatic Colorectal Cancer (mCRC) is the third most common cancer and one of the leading causes of cancer-related death worldwide and accounting for 40% to 50% of newly diagnosed patients with high mortality rates. The 5-year overall survival (OS) is very low, which is 18 to 21 months even with the advancement of chemotherapeutic treatment. Two monoclonal antibodies (MoAbs), Cetuximab and Panitumumab, which target Epidermal Growth Factor Receptor (EGFR), have been approved more recently to treat mCRC. These two MoAbs target EGFR by binding to the extracellular domain and thus, leading to the inhibition of its downstream signaling. They have proven a modest clinical benefit in pretreated patients by the use of either, alone or in combination with conventional chemotherapy. It became clear from the beginning that not all the patients with mCRC benefit from these anti-EGFR MoAbs (1). Only 10% to 20% patients truly benefit from anti-EGFR MoAbs due to the high resistance against this therapy (2,3) KRAS protein, which is encoded by KRAS gene, is an early player in many signal transduction pathways (e.g., EGFR pathway). The protein product of the normal KRAS gene performs an essential function in normal tissue signaling and the mutation of a KRAS gene is an essential step in the development of many cancers. In various retrospective studies and randomized trials, show that the presence of KRAS mutations are predictive of resistance to the anti-EGFR MoAbs treatment and associated with a bad prognosis and low survival rate (1). It has been previously shown at clinical and preclinical levels that KRAS gene mutations are an independent predictive marker of anti-EGRF MoAbs resistance. On the basis of these results, The European Union Drug Regulatory Body and The European Medicine Agency have approved the use of anti-EGRF MoAbs therapy, for only those patients exhibiting mCRC with wild-type KRAS (4). It is found that in human CRC, mutations in KRAS genes are very frequent, however, between 20% to 50% of total non responsive patients (4,5). Even the presence of wild-type KRAS does not guarantee the full benefit from anti-EGFR MoAbs therapy. In the absence of KRAS mutations, resistance to anti-EGFR MoAbs treatments may possibly be caused by the alterations in the downstream members of RAS-RAF-MAPK pathway?
Introduction
BRAF, one of the members of the three protein-serine/threonine kinases that are related to retroviral oncogenes, was discovered in 1988. Owing to prior DNA sequencing error, BRAF residue numbering changed in 2004. In the original version, residues after 32 were one number shorter than their actual position.
BRAF is major downstream effectors of KRAS and is also considered an oncogene whose activating mutations appear in about 12-18% of human colorectal cancer (6). BRAF plays a role in the regulation of mitogen-activated protein/extracellular signal-regulated kinases MAP/ERKs signaling pathway, which controls the cellular division, differentiation and secretion. Mutations in this gene can lead to different diseases including CRC.
Factors involving in B-RAF mutations and impared signaling
The activation of BRAF oncogene, inactivation of mismatch repair genes by methylation of CpG islands, and microsatellite instability (MSI) have been reported to be involved in CRC development (7). B-RAF does not require additional negative charge during activation by additional enzyme modification, since its N-region contains an activating serine site and the basal activity of BRAF is higher than its other RAF family members (8), that is why BRAF is more prone to mutations than CRAF and ARAF (9). Single amino acid substitutions can cause the activation of BRAF, but CRAF and ARAF require two mutations for their oncogenic activation, which is a very rare event to be seen (8). The most common BRAF mutation, which accounts for more than 90% of the cases of cancer involving this gene, is a glutamic acid for valine substitution at position 600 (V600E) (9). Continued expression of BRAF V600E is required for tumor growth and progression (10).
BRAF is a major contributor to many cancers. Somatic mutations in the BRAF gene have been detected in almost 50% malignant melanomas and many other cancers including CRC, ovarian and papillary thyroid carcinomas (11).
Of the oncogenic mutations in the BRAF gene, most are clustered in two regions of the kinase domain, which is responsible for maintaining the inactive catalytic conformation, the glycine rich loop and the activation segment. The proteins of BRAF oncogene with impaired kinase activity and the binding and activation of CRAF are required for ERK activation in vivo. The oncogenic BRAF proteins have been divided into three groups based on their enzymatic activity: (I) those with high enzymatic activity, they are 130-700 folds more active than the wild-type (WT) BRAF; (II) those with intermediate activity, which are 60 to 1.3 folds more active than WT BRAF; (III) those with impaired catalytic activity are 0.8 to 0.3 folds active as compared to WT BRAF (12). Activating mutations in BRAF oncogene have been reported in 10-15% CRC with the vast majority being a V600E hotspot mutation (13). V600E substitution is strongly associated with microsatellite instability (MSI+) phenotype, but is mutually exclusive with KRAS mutations (14). CIMP provides a unique opportunity to study the molecular mechanism that leads to epigenetic changes in CRC and how these changes can cause this disease (15). A strong association between CpG island methylator phenotype (CIMP) and the presence of an activated form of BRAF mutation (BRAFV600E) has been founded (16). It has also been demonstrated that sporadic microsatellite instability (MSI) occurs as a consequence of CIMP-associated MLH1 DNA hypermethylation (16,17). It has been reported that both BRAF mutations and CIMP are present in the earliest stages of colorectal neoplasia, where CIMP is present in apparently normal mucosa of patients predisposed to multiple serrated polyps (18) as well as BRAF mutations in aberrant crypt foci (19). BRAF mutations in tumors with MSI+ CIMP+ are 10 folds more frequent than tumors without these phenotypes (20), 70-80% BRAF mutation frequencies have been reported in sporadic MSI+, CIMP+ and MLH1-methylated CRC and polyps (21). The BRAF oncogene gene has been linked to MSI pathway in tumorigenesis (22).
BRAF mutation frequencies in MSI+ are much higher than MSI- tumors, and the higher frequencies have been seen in tumors showing methylation of the MLH1 promoter proximal region and in tumors with infiltrating lymphocytes (20). It has been reported in various studies that 100% of the carcinomas with BRAF mutations, methylation of hMLH1 occurred. Samowitz et al. have speculated about a fact, that MSI colorectal tumors that develop from hyperplastic polyps frequently show BRAF mutations and the methylator phenotype (CIMP), including the methylation of hMLH1 (23). According to Domingo et al, the inactivation of hMLH1 by methylation is reacted to the activation of BRAF, suggesting that specific modulation in the RAS/RAF system could occur depending on hMLH1 methylation status in CRC (24).
Koinuma K et al. reported an association between BRAF mutations and promoter methylation of the hMLH1 repair gene, where hMLH1 has been found to be altered in 80% of the cases of MSI sporadic CRC (25). BRAF mutations were reported to show prognostic significance in MSI - but not in MSI+ CRC (26). In various previous studies it has been reported that BRAF mutations in MSI-sporadic CRC are more frequently detected as compared with microsatellite -stable CRC (up to 50% vs. 12% respectively) (26).
During uncontrolled division in tumor cells, their demand for nutrients and oxygen increases, and to adapt to hypoxic environment, cells switch to anaerobic glycolysis and induction of survival factors and angiogenic growth factors such as; vascular endothelial growth factor (VEGF) (27). In hypoxia, Hypoxia-inducible factors (HIFs) are thought to play a major role in controlling the transcriptional responses (28). Mutated BRAF induces and regulates both Hypoxia-inducible Factor-1α (HIF-1α) and Hypoxia inducible Factor -2α (HIF-2α) in hypoxia (29). KRAS induces only HIF-1α. HIF-1α is thought to promote the growth of colon cancer cells, whereas; HIF-2α may restrain growth. The differential effects of KARS and BRAF mutations on the HIFs presents the unique interaction between the oncogenes and the tumor microenvironment, which may provide the phenotypic differences in mutant BRAF and KRAS CRC (29).
MoAbs action with non-BRAF mutated and BRAF mutated cells
Two monoclonal anti-bodies (MoAbs), Cetuximab and Panitumumab, which target the EGFR have been entered in clinical practice to treat mCRC, both these molecules bind to the EGFR external domain, leading to inhibition of its downstream signaling pathways (Figure 1).These include the RAS-RAF-MAPK axis, which is mainly involed in cell proliferation, and the P13K-PTEN-AKT pathway, which is involved in cell survival and motility (30).
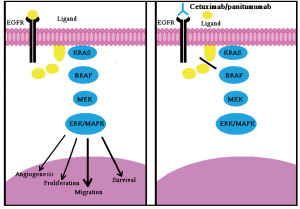
The anti-EGFR monoclonal antibody, Cetuximab, has demonstrated clinical beneifits in, and is widely used to treat, mCRC (Figure 1) (31). Notion has been acknowledged by European Medicine Agency (EMEA), which approved the use of Panitumumab or Cetuximab only in mCRC patients whose tumors display wt-KRAS (32). American Society of Clinical Oncology recommended that only those mCRC patients with wild-type KRAS be considered candidates to receive anti-EGFR therapy. The efficacy of anti-EGFR monoclonal antibodies in 60-70% of mCRC patients with wt-KRAS tumors is still limited, with response rates between 10% and 40% (33). There is a need for additional biomarkers for these patients. Interestingly the expression of the EGFR protein has not been strongly associated with clinical response to Cetuximab in CRC, although, there is limited evidence that amplification of the EGFR gene relates to objective response and other indices of clinical benefits. The relation between the increase of the EGFR gene dosage and response to Cetuximab or Panitumumab is not strong enough to allow the clinical use of this biomarker for the predictive selection of patients (34).
As proven, BRAF is the principal effectors of KRAS (35) and its oncogenic V600E mutation is mutually exclusive with KRAS mutations in CRCs (36). It has been demonstrated that the V600E mutation can also preclude responsiveness to Panitumumab or Cetuximab in mCRC patients and cellular models of CRC also, mutations in BRAF have shown impaired responsiveness to Panitumumab or Cetuximab in patients with mCRC (Figure 2A) (4). Of note, KRAS and BRAF mutations are known to be mutually exclusive in colorectal cancers (36). Patients who have mutated BRAF don’t respond to MoAbs therapy even if they present wt-KRAS, which shows that wt-BRAF is required to respond to MoAbs therapy to treat mCRC (4). Therefore, mutated BRAF tumors (approximately 10%) add algebraically to those carrying KRAS mutations (35-45%), thus further empowering the selection of patients eligible for Cetuximab/Panitumumab treatment. Of note, when considered together, the two biomarkers can identify up to 55% non-responders (4).
BRAF inhibitors
As there are no drugs currently available for the specific and direct inhibition of KRAS, but there are number of agents that are designed to inhibit the kinase activity of BRAF, which is either already clinically approved or is progressing through the pipeline of phase I and II studies (37).
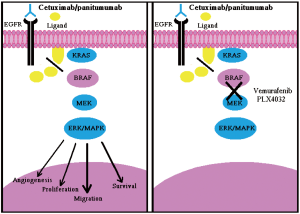
The pharmacological inhibition of the MAPK signaling cascade in cancer cells carrying constitutively active KRAS and BRAF mutants has been shown to improve anti-EGFR treatment with MoAbs. In this regard, it has been reported that treatment with the BRAF inhibitor, Sorafenib, can restore sensitivity to Cetuximab and Panitumumab of CRC cells carrying the V600E allele (38).
So, the concomitant treatment of patients with mCRC bearing BRAF-mutated tumors, with Cetuximab/Panitumumab in combination with a BRAF inhibitor, is possible and supported by a strong rationale. MoAbs activity can be restored in BRAF mutated patients by introducing BRAF inhibitor along with MoAbs therapy (Figure 2B). Recently another study has reported the preclinical characterization of Vermurafenin (RG7202;PLX4032;RO5185426), which is a first-class specific small molecule BRAFV600E inhibitor in BRAF -mutated CRC cell lines and tumor xenograft models. In the same study Vermurafenin showed the dose dependent inhibition of ERK and MEK phosphorylation, which caused the inhibition of tumor growth in BRAFV600E, bearing xenograft models and arresting of cell proliferation in BRAFV600E expressing cell lines. This shows that combination of Vemurafenib with MoAbs therapy could enhance the clinical anti tumor efficacy in CRC harboring the BRAFV600E mutation (Figure 2B) (39). It has been shown that the multikinase inhibitor, Sorafenib, might restore sensitivity to EGFR inhibitors in BRAF mutated CRC cell lines, and combining of more selective BRAF inhibitors [e.g., PLX-4032 and XL-281 can also restore the sensitivity EGFR-targeted antibodies towards BRAF mutation (4)]. The first generation of RAF inhibitors, including sorafenib, were notable for their lack of specificity and potency for RAF and these agents have shown limited efficacy in tumors with a high incidence of BRAF mutation, such as, melanoma. Novel inhibitors of the pathway with greater selectivity for BRAF and MEK are now in Phase 1 and 2 clinical trials with promising early results. To maximize the likelihood of success with these agents, clinical trials enriched with patients whose tumors possess BRAF and RAS mutations have been proposed (40). It has also been reported that AZ628, a selective and potent investigational small molecule RAF kinase inhibitor, is remarkably effective at inhibiting the growth of a specific subset of human cancer cell lines derived from melanomas, thyroid cancers, and colorectal cancers that harbor the BRAF V600 mutation (41).
Resistance to BRAF inhibitors
Clinical responses to target anticancer therapeutics are frequently confounded by de novo or acquired resistance (42). In chronic myelogenous leukemia (CML), gastrointestinal stromal tumors (GIST) and non-small cell lung cancers (NSCLCs), acquired resistance to kinase inhibitors is frequently associated with either secondary kinase domain mutations, amplification of the gene encoding the target kinase, or mutational activation of genes encoding components of alternative survival pathways (42-46). It has been shown that elevated levels of CRAF cause the acquired resistance to BRAF inhibition in the melanomas (47). It has also been shown that MAP3K8 (COT/TPL2), which is a MAPK pathway agonist, drives resistance to RAF inhibition in cell lines containing RAFV600E mutation (48).
Overcoming BRAF inhibitors’ resistance
As various targeted kinase inhibitors have demonstrated both pre-clinical and clinical activity, the application of these agents to large patient population has clearly demonstrated that while initial clinical responses can be dramatic, rapid acquisition drug resistance is a major limitation to the over therapeutic efficacy of these drugs. Therefore, one of the major challenges associated with the border use of these inhibitors is the elucidation of drug resistance mechanisms and the development of strategies to overcome or prevent resistance.
Identification of resistance mechanisms in a manner that elucidate alternative “druggable” targets may inform effective long-term treatment strategies (49). Each of these identified resistances in CLM,GIST and NSCLC, has been successfully modeled in cell culture using appropriate drug-treated cancer cell lines, indicating that such cell culture modeling can provide an effective system for identifying mechanism of acquired drug resistance that are likely to arise clinically (46,50,51).This is important because the development of strategies to overcome drug resistance, which will generally requires considerable time, first requires the identification of relevant resistance mechanisms. Therefore, the ability to anticipate clinical mechanisms of acquired resistance to targeted kinase inhibitors is likely to greatly accelerate the development of strategies to overcome drug resistance (52), and to reduce the current temporal gap between initial clinical successes and subsequent disease progression in the absence of available secondary treatment options. Anticipating the potential mechanisms of acquired resistance that could develop to the RAF inhibitors during the course of treatment can overcome this problem, as drug resistant clones from human melanoma-derived cell line harboring the V600E activating mutation that showed sensitivity to AZ628, a selective RAF kinase inhibitor. In the subset of these clones, significantly increased expression of the BRAF-related CRAF protein appeared to account for the acquisition of resistance to AZ628. But the resistant clones, which have shifted their dependency from BRAF to CRAF, acquired substantial sensitivity to the HSP90 inhibitors, Geldanamycin, which promotes the degradation of CRAF, thereby revealing a potential therapeutic strategy to overcome this resistance mechanism (47).
BRAF mutation analysis in CRC
As in July 2009, the Food and Drug Administration (FDA) approved labeling changes to two EGFR antagonists, Cetuximab and Panitumumab, stating that these agents are not recommended for the treatment of CRC harboring KRAS mutations. Thus, determination of KRAS mutation status in these tumors is critical when evaluating a patient for anti-EGFR therapy.
The American Society of Clinical Oncology (ASCO) has further recommended that all patients with metastatic colorectal cancer, for whom EGFR antagonists are being considered, should be specifically tested for KRAS mutational status at codons 12 and 13. Current guidelines in the US state, that patients with metastatic CRC being considered for EGFR-targeted therapies should be tested for KRAS and BRAF mutations (53)
Data from the CRYSTAL trial suggest that BRAF mutations are also indicative of poor prognosis and the National Comprehensive Cancer Network (NCCN), Colon Cancer Guideline Update 2010 states that testing for mutations in BRAF should occur when KRAS testing indicates KRAS wild type, to avoid exposing patients to ineffective drugs, exposure to unnecessary drug toxicities, and expedite the use of the best available alternative therapy (54). High-resolution melting (HRM) is a recently developed technique that shows great potential for scanning germline and somatic mutations (55).
Also, another recent study described HRM assay for mutation detection in EGFR exons 19-21, KRAS codon 12/13 and BRAF V600 using formalin-fixed paraffin embedded samples, which proved HRM as a rapid and sensitive method for moderate-throughput cost-effective screening of oncogene mutations in clinical samples (56).
BRAF mutations are now increasingly being investigated in metastatic colorectal cancer. KRAS mutation analysis may be considered medically necessary to predict no response to anti-EGFR monoclonal antibodies Cetuximab and Panitumumab in the treatment of metastatic, unresectable, or advanced colorectal cancer.
BRAF mutation analysis is considered investigational for all indications, including, but not limited to, predicting no response to anti-EGFR monoclonal antibodies Cetuximab and Panitumumab in the treatment of metastatic, unresectable, or advanced colorectal cancer.
KRAS and BRAF mutation analyses using PCR methodology are commercially available as laboratory-developed tests. Such tests are regulated under the Clinical Laboratory Improvement Amendments (CLIA). Premarket approval from the U.S. Food and Drug Administration (FDA) is not required when the assay is performed in a laboratory that is licensed by CLIA for high-complexity testing.
Factors affecting the efficacy of MoAbs Cetuximab In CRC patients other than BRAF, KRAS mutations
Mutated BRAF tumors (approximately 10%) add algebraically to those carrying KRAS mutations (35% to 45%), thus further empowering the selection of patients eligible for Cetuximab/Panitumab treatment. When considered together, the two markers can identify up to 55% of non responders (4). Results from other groups recently reported at the 2009 annual meeting of the American Association of Cancer Research and the American Society of Clinical Oncology confirmed these data (57).
In addition to KRAS and BRAF, the EGF receptor also activates the PI3k signaling pathway. This signaling pathway can be oncogenically deregulated either by activating mutations in the PIK3CA p110 subunit or by inactivation (often by epigenetic mechanisms) of the PTEN phosphatase. The role of deregulated PIK3CA/PTEN signaling on the response to Cetuximab and Panitumab has therefore been investigated. As in one study, it is indicated that when expression of PTEN and mutations of KRAS, BRAF and PIK3CA concomitantly ascertained up to 70% of patients with mCRC unlikely to respond to anti-EGFR therapies, can be identified (58). A gross analysis of current data regarding the impact of PIK3CA mutations and PTEN loss on response is conflicting (59-63). From the published work, it seems that PIK3CA mutations are in fact associated with the resistance, although, this correlation is nowhere close to that observed for KRAS or BRAF. However, most of the authors agree that PTEN inactivation is a negative predictor of response (59,64).
As KRAS and BRAF mutations are exclusive, but the mutations of PIK3CA or inactivation of PTEN can coexist [i.e., they can occur in the same tumor containing KRAS/BRAF mutations (3).], which makes it difficult to find the individual contribution of PIK3CA mutations and PTEN inactivation to the resistance against MoAbs therapy other than KRAS and BRAF mutations. It has also been shown that PIK3CA mutations located in exon 9 and 20 hotspots exert different biochemical and oncologic properties and are differently activated by KRAS (65). So, it is convincible that both PIK3CA mutations and PTEN inactivation have a little contribution of resistance against Cetuximab and Panitumumab therapy due to co-occurrence of PTEN expression and PIK3CA mutations with KRAS and BRAF mutations and different oncogentic properties of different PIK3CA mutations, so for definite conclusions more research work and analyzing of large cohorts of patients are needed to become useful to further analyze the eligible patients to treat with MoAbs therapy. However, these two markers are not yet ready to use clinically. Other possibilities can be the occurrence of alterations in other key elements of the EGFR-dependent signal cascade (e.g., AKT1 or MEK- MAPK), as in preclinical studies, inhibition of the MEK kinase effectively and specifically inhibits the growth of human tumor cells lines harboring activating BRAF mutations (66) and genetic alternation in tyrosine kinase receptors other than EGFR, providing an alternate pathway of survival and/or proliferation.
The Relationship between the increase EGFR gene dosage and response to Cetuximab or Panitumumab is not strong enough to allow the clinical use of this biomarker for the predictive selection of patients (34).
Secondary resistance to MoAbs therapies in mCRC patients is another cause of ineffectiveness, therefore, it is important to identify the possible mechanism causing secondary resistance. As has been mentioned in a clinical data, the response is transient, even in the KRAS and BRAF wild type tumors, and only last for 1 to 1.5 years (67).
The somatic knocking-out or knocking-in of individual alleles in normal or neoplastic cells is a new generation of cell tumor progression models, which has been developed recently. Generation of paired cell lines which closely recapitulate the occurrence of cancer mutations in individual patients as a result of targeting the endogenous loci for mutation or correction (68,69). It has been shown that the growth of human tumor cell lines harboring activating BRAF mutations can be inhibited by effective and specific inhibition of MEK kinase (66).
Role of ethnicity, gender and smoking in BRAF mutated mCRC
The link of BRAF and KRAS mutations with ethnicity has been reported. In Chinese and Caucasians BRAF mutations were reported to be associated with advance disease stages and worse survival of papillary thyroid microcarcinoma (70,71), but not in Japanese (54). A study from Australia showed that people of southern Europe origin had a lower risk of BRAF mutation then those of Anglo-Celtic origin (72). BRAF mutations were detected in about 45% of the high microsatellite instability (MSH-H) tumors and in about 10% of the microsatellite stable (MSS) tumors in Caucasians (73). In African Americans, distinct BRAF mutation has been reported, with 23% in MSI tumors and 0% in non-MSI tumors (74). These findings show the relation and importance of investigation of BRAF mutations with different ethnic groups.
In colorectal cancers, BRAF and PIK3CA (but not KRAS, APC, or TP53) mutations display a gender bias at higher frequencies in females (75,76). This suggests that tumors with BRAF somatic mutations arise from a different pathway in women. As one study has reported that exposure to estrogen in women protects against MSI, whereas, the lack of estrogen in aged females increases the risk of instability (77). Use of Hormone Replacement Therapy (HRT) significantly reduces the risk of colon cancer in postmenopausal females (78).This shows that the lack of female hormones contributes in the development of various cancers including colon cancer, which suggests that it could be hypothesized that female patients with mCRC might be less likely to benefit from treatment with EGFR-targeted MoAbs. However, available clinical data do not support this hypothesis (79,80).
Smoking is also associated with mCRC caused by BRAF mutations but it is not as strongly associated as gender, though females are twice likely to have a tumor with BRAF mutation, but it is not strongly associated with smoking, as men who smoke are at higher risk of mCRC with BRAF mutations (81).
Conclusions
In addition to KRAS analysis, BRAF analysis should be done to rule out the BRAF mutations, especially in the developing countries like China, where BRAF testing is not common (other than few metropolitan areas), to avoid the unnecessary cytotoxicity, for selecting patients who would respond to the therapies and as the shocking costs of these targeted therapies, so the selection of patients is the key role to their economic sustainability. And besides using Anti-EGFR MoAbs, other alternative therapies should also be considered. As current data suggests that evaluation of not only KRAS or BRAF but also P1k3CA/PTEN alterations could be useful for selecting patients with mCRC who are unlikely to respond to anti Anti-EGFR-MoAbs. Genetic manipulation techniques can be applied to cellular models, one can envisage developing in vitro tools to prospectively find new sensitivity resistance biomarkers that can then be confirmed in patients and even be used to screen for rationale drug combinations to reverse resistance. To restore the sensitivity of MoAbs, they could be administered along with BRAF inhibitors and at the same time new ways should be found out in order to reduce the resistance to the BRAF inhibitors, further understanding of the molecular mechanisms to discover new alternative therapies and tests for non-responding patients would be helpful.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Saridaki Z, Tzardi M, Papadaki C, et al. Impact of KRAS, BRAF, PIK3CA mutations, PTEN, AREG, EREG expression and skin rash in >/= 2 line cetuximab-based therapy of colorectal cancer patients. PLoS One 2011;6:e15980.
- Mao C, Liao RY, Qiu LX, et al. BRAF V600E mutation and resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer: a meta-analysis. Mol Biol Rep 2011;38:2219-23.
- Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol 2010;28:1254-61.
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26:5705-12.
- Shen H, Yuan Y, Hu HG, et al. Clinical significance of K-ras and BRAF mutations in Chinese colorectal cancer patients. World J Gastroenterol 2011;17:809-16.
- Makrodouli E, Oikonomou E, Koc M, et al. BRAF and RAS oncogenes regulate Rho GTPase pathways to mediate migration and invasion properties in human colon cancer cells: a comparative study. Mol Cancer 2011;10:118.
- Rasuck CG, Leite SM, Komatsuzaki F, et al. Association between methylation in mismatch repair genes, V600E BRAF mutation and microsatellite instability in colorectal cancer patients. Mol Biol Rep 2012;39:2553-60.
- Emuss V, Garnett M, Mason C, et al. Mutations of C-RAF are rare in human cancer because C-RAF has a low basal kinase activity compared with B-RAF. Cancer Res 2005;65:9719-26.
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54.
- Hoeflich KP, Gray DC, Eby MT, et al. Oncogenic BRAF is required for tumor growth and maintenance in melanoma models. Cancer Res 2006;66:999-1006.
- Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev 2007;17:31-9.
- Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004;116: 855-67.
- Yuen ST, Davies H, Chan TL, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res 2002;62:6451-5.
- Oliveira C, Pinto M, Duval A, et al. BRAF mutations characterize colon but not gastric cancer with mismatch repair deficiency. Oncogene 2003;22:9192-6.
- Schuebel K, Chen W, Baylin SB. CIMPle origin for promoter hypermethylation in colorectal cancer? Nat Genet 2006;38:738-40.
- Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787-93.
- Hinoue T, Weisenberger DJ, Pan F, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One 2009;4:e8357.
- Minoo P, Baker K, Goswami R, et al. Extensive DNA methylation in normal colorectal mucosa in hyperplastic polyposis. Gut 2006;55:1467-74.
- Rosenberg DW, Yang S, Pleau DC, et al. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer Res 2007;67:3551-4.
- Li WQ, Kawakami K, Ruszkiewicz A, et al. BRAF mutations are associated with distinctive clinical, pathological and molecular features of colorectal cancer independently of microsatellite instability status. Mol Cancer 2006;5:2.
- Yang S, Farraye FA, Mack C, et al. BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol 2004;28:1452-9.
- Maestro ML, Vidaurreta M, Sanz-Casla MT, et al. Role of the BRAF mutations in the microsatellite instability genetic pathway in sporadic colorectal cancer. Ann Surg Oncol 2007;14:1229-36.
- Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005;129:837-45.
- Domingo E, Espín E, Armengol M, et al. Activated BRAF targets proximal colon tumors with mismatch repair deficiency and MLH1 inactivation. Genes Chromosomes Cancer 2004;39:138-42.
- Koinuma K, Shitoh K, Miyakura Y, et al. Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas. Int J Cancer 2004;108:237-42.
- Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res 2005;65:6063-9.
- Shweiki D, Itin A, Soffer D, et al. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature 1992;359:843-5.
- Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Pflugers Arch 2005;450:363-71.
- Kikuchi H, Pino MS, Zeng M, et al. Oncogenic KRAS and BRAF differentially regulate hypoxia-inducible factor-1alpha and -2alpha in colon cancer. Cancer Res 2009;69:8499-506.
- Baselga J. The EGFR as a target for anticancer therapy--focus on cetuximab. Eur J Cancer 2001;37:S16-22.
- Van Cutsem E, Vervenne WL, Bennouna J, et al. Phase III trial of bevacizumab in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. J Clin Oncol 2009;27:2231-7.
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34.
- Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009;27:2091-6.
- Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol 2005;6:279-86.
- Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J 2000;19:5429-39.
- Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002;418:934.
- Garber K. Trial offers early test case for personalized medicine. J Natl Cancer Inst 2009;101:136-8.
- Di Nicolantonio F, Martini M, Molinari F, et al. BRAF V600E confers resistance to cetuximab or panitumab metastatic colorectal cancer. Eur J Cancer 2008;44:6.
- Yang H, Higgins B, Kolinsky K, et al. Antitumor activity of BRAF inhibitor vemurafenib in preclinical models of BRAF-mutant colorectal cancer. Cancer Res 2012;72:779-89.
- Pratilas CA, Solit DB. Therapeutic strategies for targeting BRAF in human cancer. Rev Recent Clin Trials 2007;2:121-34.
- McDermott U, Sharma SV, Dowell L, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A 2007;104:19936-41.
- Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol 2006;24:4764-74.
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73.
- Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 2001;293:876-80.
- Donato NJ, Wu JY, Stapley J, et al. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood 2003;101:690-8.
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43.
- Montagut C, Sharma SV, Shioda T, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res 2008;68:4853-61.
- Johannessen CM, Boehm JS, Kim SY, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 2010;468:968-72.
- Daub H, Specht K, Ullrich A. Strategies to overcome resistance to targeted protein kinase inhibitors. Nat Rev Drug Discov 2004;3:1001-10.
- Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell 2003;112:831-43.
- Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest 2006;116:2695-706.
- Azam M, Daley GQ. Anticipating clinical resistance to target-directed agents : the BCR-ABL paradigm. Mol Diagn Ther 2006;10:67-76.
- National Comprehensive Cancer Network Guidelines for Colon and Rectal Cancer 2010: v.1.
- Ito Y, Yoshida H, Maruo R, et al. BRAF mutation in papillary thyroid carcinoma in a Japanese population: its lack of correlation with high-risk clinicopathological features and disease-free survival of patients. Endocr J 2009;56:89-97.
- Taylor CF. Mutation scanning using high-resolution melting. Biochem Soc Trans 2009;37:433-7.
- Borràs E, Jurado I, Hernan I, et al. Clinical pharmacogenomic testing of KRAS, BRAF and EGFR mutations by high resolution melting analysis and ultra-deep pyrosequencing. BMC Cancer 2011;11:406.
- Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 2009;27:5924-30.
- Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, et al. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS One 2009;4:e7287.
- Frattini M, Saletti P, Romagnani E, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer 2007;97:1139-45.
- Genome Web Daily News: FDA adds KRAS testing info to Vectibix,Erbitux Labels(press Release,GEnome Web News). Available online: http://www.genomeweb.com/dxpgx/fda-adds-kras-testing-info-vectibix-erbitux-labels
- Jhawer M, Goel S, Wilson AJ, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res 2008;68:1953-61.
- Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 2009;69:1851-7.
- Prenen H, De Schutter J, Jacobs B, et al. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res 2009;15:3184-8.
- Perrone F, Lampis A, Orsenigo M, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol 2009;20:84-90.
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A 2008;105:2652-7.
- Solit DB, Garraway LA, Pratilas CA, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature 2006;439:358-62.
- Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337-45.
- Rago C, Vogelstein B, Bunz F. Genetic knockouts and knockins in human somatic cells. Nat Protoc 2007;2:2734-46.
- Di Nicolantonio F, Arena S, Gallicchio M, et al. Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proc Natl Acad Sci U S A 2008;105:20864-9.
- Elisei R, Ugolini C, Viola D, et al. BRAF(V600E) mutation and outcome of patients with papillary thyroid carcinoma: a 15-year median follow-up study. J Clin Endocrinol Metab 2008;93:3943-9.
- Lee X, Gao M, Ji Y, et al. Analysis of differential BRAF(V600E) mutational status in high aggressive papillary thyroid microcarcinoma. Ann Surg Oncol 2009;16:240-5.
- English DR, Young JP, Simpson JA, et al. Ethnicity and risk for colorectal cancers showing somatic BRAF V600E mutation or CpG island methylator phenotype. Cancer Epidemiol Biomarkers Prev 2008;17:1774-80.
- Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 2009;58:90-6.
- Kumar K, Brim H, Giardiello F, et al. Distinct BRAF (V600E) and KRAS mutations in high microsatellite instability sporadic colorectal cancer in African Americans. Clin Cancer Res 2009;15:1155-61.
- Barault L, Veyrie N, Jooste V, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer 2008;122:2255-9.
- Benvenuti S, Frattini M, Arena S, et al. PIK3CA cancer mutations display gender and tissue specificity patterns. Hum Mutat 2008;29:284-8.
- Slattery ML, Potter JD, Curtin K, et al. Estrogens reduce and withdrawal of estrogens increase risk of microsatellite instability-positive colon cancer. Cancer Res 2001;61:126-30.
- Newcomb PA, Storer BE. Postmenopausal hormone use and risk of large-bowel cancer. J Natl Cancer Inst 1995;87:1067-71.
- Tol J, Koopman M, Cats A, et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med 2009;360:563-72.
- Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009;360:1408-17.
- Rozek LS, Herron CM, Greenson JK, et al. Smoking, gender, and ethnicity predict somatic BRAF mutations in colorectal cancer. Cancer Epidemiol Biomarkers Prev 2010;19:838-43.


