
Upregulation of ERCC6L is associated with tumor progression and unfavorable prognosis in hepatocellular carcinoma
Introduction
Despite the progress achieved in the precaution, diagnosis and treatment for hepatocellular carcinoma (HCC), its incidence and mortality remain high in the world, with over 841,000 new cases and 781,000 HCC-related deaths in 2018 (1,2). The still unresolved clinical problems in terms of its distinct characteristics (such as insidious onset, frequent relapse and poor prognosis) pose great challenges for clinicians and researchers (3). In the context of precision medicine, deeper understanding of the biological features and molecular pathogenesis of HCC will contribute to improvements in the early diagnosis, targeted therapy and long-term outcome for HCC patients (4).
ERCC6L (excision repair cross-complementation group 6-like), also known as PICH (Plk1-interacting checkpoint helicase), belongs to the family of SNF2 helicase-like ATPases, which are closely related to cell mitosis and remodeling of chromatin structures (5,6). Previous studies have reported that knocking out ERCC6L in human or animal (such as chicken and mouse) cells can lead to obvious chromosomal abnormities, DNA damage, embryonic lethality, apoptosis, and TP53 activation (7,8). In view of the crucial role of ERCC6L in preserving chromosomal integrity in fast-growing mammalian cells, it is tempting to explore the correlation between ERCC6L and tumorigenesis (8). In the last few years, ERCC6L dysregulation and its role in several cancers have been studied. For example, Pu et al. demonstrated that overexpression of ERCC6L in breast and kidney cancers correlated with the progression of disease and poorer survival of cancer patients (9). Huang et al. showed that silencing ERCC6L in cancer cells (such as liver, breast, colorectal and kidney cancers) resulted in cell cycle arrest, proliferation inhibition, invasion reduction and apoptosis (5,7,10-12). The above studies undoubtedly indicate the key implications of ERCC6L dysregulation in tumorigenesis. Nevertheless, its expression and role in HCC remain to be investigated.
In this study, utilizing public data from Human Protein Atlas (HPA) and The Cancer Genome Atlas (TCGA), ERCC6L upregulation in HCC and its clinical significance were identified. In addition, the mechanisms and biological roles underlying ERCC6L dysregulation in HCC were also preliminarily elucidated by performing comprehensive bioinformatics analyses. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-192).
Methods
Data mining in the TCGA and HPA databases
According to the inclusion criteria described previously, the level-3 data (sequencing and clinical data) of HCC patients (n=298) undergoing surgical resection plus the sequencing data of normal controls (n=50) in the TCGA dataset were ultimately included in this study (13). The raw counts of RNA-seq data were normalized by TMM methods implemented in R package “edgeR” and then log2-transformed. In addition, several online analysis platforms based on TCGA data (including UALCAN, GEPIA, cBioPortal and UCSC-Xena) were utilized for reviewing ERCC6L expression across TCGA cancers, assessing copy number alterations (CNA) and DNA methylation of ERCC6L, exploring the potential coexpression of PLK1 (polo-like kinase 1) and ERCC6L, and evaluating the impact of ERCC6L dysregulation on TP53 mutations in HCC (14-17). In particular, the Tumor Immune Estimation Resource (TIMER), an online platform for the systematic estimation of tumor-infiltrating immune cells according to the gene expression profiles of TCGA datasets, was used to determine the impact of tumor purity and immune infiltrates on the ERCC6L expression level and its prognostic value in HCC (18). Furthermore, the immunohistochemistry (IHC) data of ERCC6L (antibody: HPA050492) in normal liver and HCC tissues were obtained from the HPA database to evaluate ERCC6L protein levels (19).
Predicting the upstream miRNAs of ERCC6L
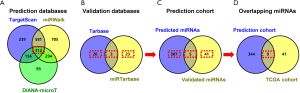
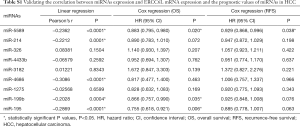
Of the 298 HCC patients, 296 samples had intact miRNA-seq data. The differentially expressed miRNAs in HCC tissues (n=296) compared to normal controls (n=50) were identified by the R package “edgeR”. A fold change >2 with a P value <0.05 was regarded as the threshold. Thus, the downregulated miRNAs in HCC were included in the TCGA cohort. In addition, the potential regulatory miRNAs of ERCC6L were obtained by three prediction databases (DIANA-microT, TargetScan v7.2 and miRWalk) and two validation databases (TarBase v.8 and miRTarBase) (20-24). The common miRNAs predicted by the three prediction databases plus all validated miRNAs in the two validation databases were considered the prediction cohort. Moreover, the overlapping miRNAs between the TCGA cohort and prediction cohort were considered underlying upstream miRNAs in HCC. Finally, the correlations between miRNA expression and ERCC6L mRNA expression and their prognostic values were verified by linear regression analysis and survival analyses.
Gene set enrichment analysis (GSEA)
GSEA using “c2.cp.kegg.v6.0.symbols.gmt” as a reference was used to recognize the possible biological pathways relevant to ERCC6L dysregulation in HCC. The gene sets with both P values and q-values less than 0.05 were regarded as significantly enriched, and their normalized enrichment scores were computed.
Construction of predictive nomograms
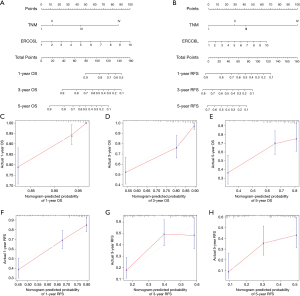
To facilitate prognosis evaluation of HCC patients, the visual, convenient and effective nomograms were constructed using the R package “rms”. The independent prognostic factors for overall survival (OS) or recurrence-free survival (RFS) prediction in HCC were determined by multivariate Cox analyses and included in the nomograms. Then, the predictive power of the nomogram was systematically assessed by concordance index (C-index) and calibration curves. The calibration curves visualized the consistency between the nomogram-predicted probability of 1-, 3- and 5-year OS (or RFS) and the actual probability of 1-, 3- and 5-year OS (or RFS). The discrimination ability of the nomogram was mathematically evaluated by calculating the C-index.
Statistical analyses
Student’s t-test or Mann-Whitney test was performed to assess gene expression levels between different groups. To measure the discrimination ability of ERCC6L for HCC, a receiver operating characteristic (ROC) analysis was utilized. X-tile 3.6.1 software was used for calculating the optimal threshold to divide patients into high- and low-expression groups. Pearson’s χ2 test was used to assess the clinical correlations of ERCC6L dysregulation in HCC. Survival analyses for OS and RFS were conducted by joint use of KM analysis and Cox regression analysis. Statistical analyses were performed by R-3.4.2 and SPSS-22.0, and a P value <0.05 was deemed as statistical significance.
Results
ERCC6L upregulation was identified in HCC tissues
Consistent with the ERCC6L upregulation generally observed in most cancers, ERCC6L was significantly upregulated at the mRNA level in HCC tissues compared to normal controls (P<0.0001) (Figure 1A,B). By performing ROC analysis, the excellent discrimination ability of ERCC6L mRNA for HCC was further identified (AUC =0.9518, P<0.0001) (Figure 1C). In addition, we found that ERCC6L expression was positively correlated with tumor purity in HCC (r=0.171, P=0.00139), which partly indicated that ERCC6L was indeed upregulated in hepatoma cells (Figure S1A). Highly consistent with its dysregulation at the mRNA level, upregulation of ERCC6L protein in HCC was also verified according to the IHC data of ERCC6L from the HPA database. Different from the ERCC6L staining level in three normal liver samples (not detected), moderate (7/10) or low (3/10) ERCC6L staining levels were usually observed in HCC tissues, in which staining was mainly localized in the cytoplasm and nucleus (Figure 1D,E).
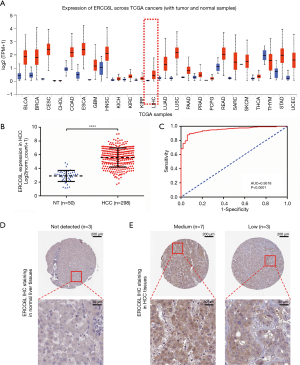
ERCC6L upregulation correlated with tumor progression and unfavorable prognosis of HCC patients
To study the clinical implications of ERCC6L dysregulation, high and low expression groups were firstly divided among the 298 patients according to the optimal cutoff value of OS. As shown in Table 1, ERCC6L upregulation was significantly related to female sex, vascular invasion (VI), elevated alpha fetoprotein (AFP), advanced histologic grade and TNM stage, and clinical outcomes (all P<0.05), and no significant correlation was found with other characteristics.
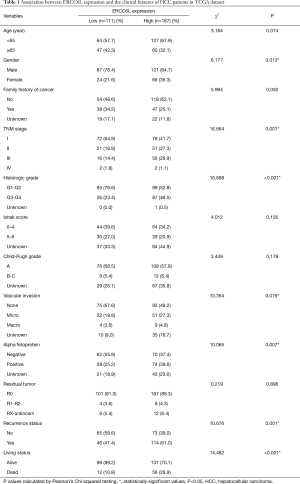
Full table
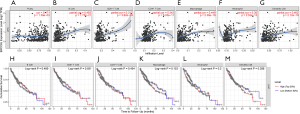
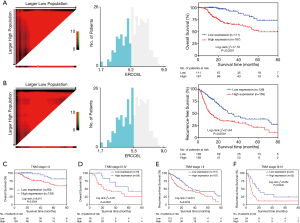
In addition, by performing KM survival analyses, worse outcomes (including OS and RFS) were observed in HCC patients with high ERCC6L expression (all P<0.05) (Figure 2A,B). Moreover, univariate Cox regression analysis indicated that ERCC6L, VI and TNM stage were related to the prognosis of HCC patients (all P<0.05). And ERCC6L upregulation was subsequently confirmed as an independent predictive factor for the poorer OS [HR =2.549 (1.327, 4.895), P=0.005] and RFS [HR =1.841 (1.324, 2.560), P<0.001] of HCC patients by multivariate analyses (Tables 2,3). In view of the significant correlation between ERCC6L expression and TNM stage, subgroup analyses were further conducted and confirmed that the prognostic value of ERCC6L did not rely on TNM stage in HCC patients (all P<0.05) (Figure 2C,D,E,F). Furthermore, the moderate correlations between ERCC6L expression and six immune cell infiltrates (B cells, CD4+T cells, CD8+T cells, neutrophils, macrophages, and dendritic cells) in HCC tissues attracted our attention and prompted us to determine the impact of immune cell infiltrates on the prognostic value of ERCC6L in HCC (Figure S1B,C,D,E,F,G) . By performing KM survival analyses and multivariate Cox regression on the TIMER online platform, we found that the six immune cell infiltrates were not related to the survival of HCC patients (Figure S1H,I,J,K,L,M) and that ERCC6L upregulation was still an independent prognostic factor for HCC patients [HR =2.300 (1.411, 3.749), P=0.001] after comprehensively adjusting for patient clinical features (including age, gender, stage and race), tumor purity and immune cell infiltrates.
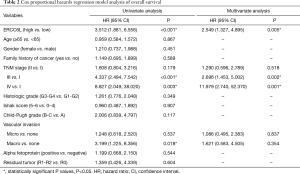
Full table
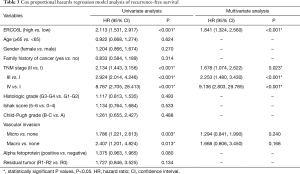
Full table
Based on the multivariate Cox regression analyses, a novel genomic-clinicopathologic nomogram integrating ERCC6L expression and TNM stage was constructed to better evaluate the prognostic and clinical application value of ERCC6L for HCC patients undergoing hepatic resection. First, calibration plots showed good consistency between the nomogram-predicted probability of survival (including OS and RFS) and the actual rate of survival. Besides, the C-index of the combined model [0.774 (0.722–0.826)] for OS prediction was clearly better than that of the TNM stage [0.698 (0.634–0.762)] (P=0.009). The C-index for RFS prediction was also improved from 0.649 (0.608–0.690) for the TNM staging system to 0.685 (0.647–0.723) for the combined model, although there was no significant difference (P=0.070) (Figure 3).
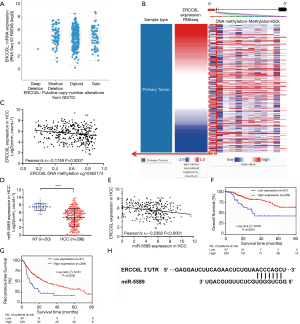
DNA hypomethylation and miR-5589 downregulation in HCC might result in ERCC6L overexpression
The genetic and epigenetic changes of ERCC6L in HCC were analyzed to explore the mechanisms underlying its dysregulation. First, the CNA of ERCC6L in HCC was evaluated based on the cBioPortal platform, and no significant association between ERCC6L upregulation and CNA was found (Figure 4A). In addition, by generating heatmaps on the UCSC-Xena platform, we observed a negative correlation between ERCC6L expression and DNA methylation. Linear regression analyses indicated that ERCC6L mRNA expression was negatively correlated with the methylation level of its specific CpG site (cg10462174, r=−0.1749, P=0.0007), which was located in the promoter of the ERCC6L gene (Figure 4B,C). Furthermore, after strict screening according to the process mentioned above (Figure S2 and Table S1), miR-5589 was identified as a potential upstream miRNA of ERCC6L. MiR-5589 was not only significantly downregulated in HCC compared to normal controls (P<0.0001), but was also significantly negatively correlated with ERCC6L mRNA level (r=−0.2362, P<0.0001) (Figure 4D,E). Importantly, survival analyses suggested that miR-5589 downregulation was related to the unfavorable prognoses of HCC patients (all P<0.05) (Figure 4F,G). But the prognostic value of ERCC6L was independent of miR-5589, as revealed by multivariate Cox regression analyses (all P<0.05) (Table S2). The putative binding site of miR-5589 on the ERCC6L 3'UTR is displayed in Figure 4H.
Full table

Full table
Biological roles underlying ERCC6L upregulation in HCC
In GSEA, eight pathways associated with ERCC6L dysregulation in HCC were found to be enriched, namely “cell cycle”, “homologous recombination”, “p53”, “DNA replication”, “mismatch repair”, “excision repair”, “nucleotide excision repair” and “spliceosome” pathways (all P<0.05) (Figure 5A,B,C,D,E,F,G,H).
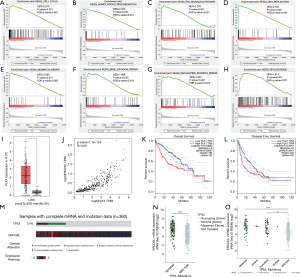
As a key interaction partner of ERCC6L, PLK1 was also upregulated in HCC tissues (P<0.05), which was positively related to ERCC6L expression (r=0.89, P<0.05) (Figure 5I,J). In addition, PLK1 upregulation correlated with worse survival outcomes in HCC patients (all P<0.05) (Figure 5K,L).
Given the results of GSEA in terms of the “p53 signaling pathway”, we attempted to analyze the potential impact of ERCC6L dysregulation on TP53 mutation using the cBioPortal platform. We surprisingly found that ERCC6L expression in the TP53 mutated groups were higher than that in the TP53 wild-type group (P<0.05), suggesting that ERCC6L upregulation might be associated with the high rate of TP53 mutation in HCC (31%, n=360) (Figure 5M,N,O).
Discussion
Considering the phenotypic heterogeneity of HCC, exploring its complicated genetic alterations and identifying potential biomarkers is crucial for molecular subtyping, early diagnosis, personalized therapy and prognosis evaluation of HCC patients. With the accumulation of evidence suggesting the involvement of ERCC6L dysregulation in several cancers, we investigated the previously unknown expression and role of ERCC6L in HCC. In line with the expression profiles of ERCC6L in most cancers, the significant upregulation of ERCC6L mRNA in HCC tissues was identified in this study, which showed good discrimination ability for HCC compared to normal liver tissues by ROC assessment. Consistent with the RNA data, ERCC6L upregulation at the protein level in HCC was further confirmed according to public IHC data. Importantly, a recent study published by Zhang et al. also demonstrated that ERCC6L protein was highly expressed in HCC tissues based on IHC results of 90 paired clinical specimens (12). The significant ERCC6L upregulation in HCC tissues suggests that it could be a valuable biomarker for HCC molecular diagnosis, and ERCC6L expression in the serum of HCC patients and its value as a noninvasive marker for HCC diagnosis (such as AFP) are worthy of future study (25).
In addition, the correlation of ERCC6L upregulation with clinicopathological characteristics (such as AFP, VI, advanced histologic grade, TNM stage and survival) in HCC strongly indicated its oncogenic role and close involvement in the development of HCC. By comprehensive survival analyses, the independent predictive value of ERCC6L was confirmed with regard to the OS and RFS of HCC patients. Relatively in line with our results, Zhang et al. reported that ERCC6L upregulation was associated with larger tumor size and worse survival in 90 HCC patients, while no correlation was found between ERCC6L upregulation and other clinicopathological features (such as AFP, VI, tumor differentiation and TNM stage) (12). We speculate that the slight differences between Zhang et al.’s study and our study may be caused by their certain limitations, including small sample size (n=90), insufficient sensitivity and discrimination of semiquantitative histochemistry score for evaluating ERCC6L expression, and improper cut-off value of ERCC6L expression for grouping (low/high). Additional larger-scale and better designed studies are warranted to validate the hypothesis in the future. Furthermore, by constructing a nomogram integrating ERCC6L expression and TNM stage that were the independent risk factors for the survival of HCC patients, the superior predictive ability of the combined model undoubtedly further supported the excellent prognostic value of ERCC6L for predicting the survival outcomes of HCC patients. Considering the still unsatisfactory predictive power of existing clinical staging systems (including the TNM staging system), ERCC6L could be a promising biomarker to establish more precise genomic-clinicopathologic staging system for HCC patients in the future, which may improve the prognosis evaluation and pre- or postoperative decision-making for HCC patients (26,27).
Genetic and epigenetic alterations of ERCC6L in HCC were also analyzed to explore the potential mechanisms underlying its upregulation. First, no significant correlation between CNA and ERCC6L expression in HCC was observed in this study. In addition, it has been confirmed that DNA methylation regulates the expression and repression of genes, and promoter-specific hypomethylation is responsible for chromosomal instability and oncogene activation in cancers (28). Hence, the impact of DNA hypomethylation on ERCC6L expression in HCC was explored in this study. By generating a heatmap and performing linear regression analysis, we found a negative association between ERCC6L expression and the methylation level of one specific CpG site in its promoter (cg10462174), which might contribute to ERCC6L upregulation in HCC. In addition, given the regulatory role of miRNAs in mRNA degradation, the possible miRNAs controlling ERCC6L expression were further investigated in this study (29). By integrated analyses, miR-5589 was identified as a potential upstream miRNA of ERCC6L in HCC, in view of the miR-5589 downregulation in HCC and its negative correlation with ERCC6L mRNA level, the putative binding site of ERCC6L by miR-5589 and the significant prognostic value of miR-5589 in HCC. In line with our results, miR-5589 downregulation in several cancers (including HCC) has been reported in previous studies (30,31). Thus, our findings initially demonstrated that both ERCC6L promoter hypomethylation and miR-5589 downregulation could be the mechanisms underlying ERCC6L upregulation in HCC. Additional basic experiments are necessary to further verify them in the future.
In light of the close involvement of ERCC6L in HCC, its potential biological functions in the development of HCC were explored in this study by enrichment analysis, which suggested that ERCC6L was implicated in the “DNA replication”, “cell cycle”, “p53” and DNA repair-related pathways. Consistent with this, ERCC6L is a known key interaction partner of PLK1 (a key controller of M phase and mitotic progression) and a DNA translocase that is responsible for the fidelity and rapid sister chromatid segregation during mitosis via proper chromosome compaction and timely resolution of ultrafine DNA bridges (8,32,33). ERCC6L deletion in mammalian cells can lead to cell cycle arrest, DNA damage, chromosomal abnormities, TP53 activation and apoptosis (8,33). Importantly, upregulation of ERCC6L has been identified in several cancers and is related to cancer progression and poorer prognosis of patients (5,7,9-11). Huang et al. found that ERCC6L was indispensable for the chromosome stability of cancer cells, and silencing ERCC6L could lead to apoptosis induction and proliferation inhibition of triple-negative breast cancer cells in vivo and in vitro by inducing the formation of chromatin bridges and mitotic catastrophe (7). Cytoplasmic and nuclear immunostaining of ERCC6L in HCC cells supports previous findings that ERCC6L is mostly located in the cytoplasm during interphase and moves to chromatin in prometaphase (8). Thus, there is a possibility that ERCC6L upregulation in HCC may contribute to the maintenance of chromosomal integrity and accelerated cell cycle progression, finally leading to unrestricted cell proliferation and tumor progression. Strongly consistent with our hypothesis, Zhang et al. demonstrated that ERCC6L upregulation in HCC could accelerate the G1/S transition of HCC cells and promote tumor proliferation via in vitro and in vivo experiments (12). In addition, the dynamic localization of ERCC6L has been reported to be regulated by PLK1, and their colocalization at kinetochores coordinately maintains chromosome architecture and genomic stability during mitosis (34,35). In line with this, PLK1 upregulation and coexpression of PLK1 and ERCC6L in HCC were also observed in this study. Given that both ERCC6L and PLK1 are prognostic indicators for HCC patients, the ERCC6L-PLK1 complex may play a crucial role during HCC progression. Furthermore, in view of the emerging evidence suggesting the correlation between ERCC6L and TP53, it is tempting to explore their relationship in HCC (8). TP53 has been identified as a classical tumor suppressor and a high rate of TP53 mutation is common in cancers (including HCC, approximately 31% in the TCGA dataset) (36). It has been demonstrated that TP53 mutations not only cause TP53 to lose its anticancer role, but also serve to protect cancer cells from DNA damage and facilitate their proliferation and tumor progression (37). In line with this, previous study showed that TP53 mutation in HCC was significantly associated with worse clinical outcomes in HCC patients (13). Considering the high level of ERCC6L in the TP53 mutated group in HCC, it is possible that ERCC6L may help to preserve the stabilization of TP53 genetic mutations after oncogenic transformation, thereby promoting tumor progression in HCC. Interestingly, Zhang et al. also noted a potential correlation between ERCC6L and the p53 pathway in HCC and attempted to shed light on their relationship from the perspective of the negative regulatory effects of ERCC6L upregulation on p53 expression levels in HCC (12). The above evidence strongly indicates that ERCC6L may be a promising therapeutic target in HCC, which deserves further researches in the future.
In conclusion, ERCC6L upregulation in HCC and its correlations with tumor progression and worse clinical prognosis of HCC patients were demonstrated in this study. ERCC6L promoter hypomethylation and miR-5589 downregulation might result in ERCC6L overexpression in HCC. ERCC6L upregulation in HCC was significantly related to the “p53”, “DNA replication” and “cell cycle” pathways, which preliminarily shed light on its biological functions in HCC. Further studies are urgently needed to elucidate the exact role of ERCC6L in HCC.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant number: 81570079).
Footnote
Reporting Checklist: The authors present the study in accordance with the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-192
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-192). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol 2020;72:250-61. [Crossref] [PubMed]
- Nahon P, Vibert E, Nault JC, et al. Optimizing curative management of hepatocellular carcinoma. Liver Int 2020;40 Suppl 1:109-15. [Crossref] [PubMed]
- Rebouissou S, Nault JC. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol 2020;72:215-29. [Crossref] [PubMed]
- Zhang G, Yu Z, Fu S, et al. ERCC6L that is up-regulated in high grade of renal cell carcinoma enhances cell viability in vitro and promotes tumor growth in vivo potentially through modulating MAPK signalling pathway. Cancer Gene Ther 2019;26:323-33. [Crossref] [PubMed]
- Ryan DP, Owen-Hughes T. Snf2-family proteins: chromatin remodellers for any occasion. Curr Opin Chem Biol 2011;15:649-56. [Crossref] [PubMed]
- Huang Y, Li W, Yan W, et al. Loss of PICH promotes chromosome instability and cell death in triple-negative breast cancer. Cell Death Dis 2019;10:428. [Crossref] [PubMed]
- Albers E, Sbroggio M, Pladevall-Morera D, et al. Loss of PICH Results in Chromosomal Instability, p53 Activation, and Embryonic Lethality. Cell Rep 2018;24:3274-84. [Crossref] [PubMed]
- Pu SY, Yu Q, Wu H, et al. ERCC6L, a DNA helicase, is involved in cell proliferation and associated with survival and progress in breast and kidney cancers. Oncotarget 2017;8:42116-24. [Crossref] [PubMed]
- Xie Y, Yu J, Wang F, et al. ERCC6L promotes cell growth and invasion in human colorectal cancer. Oncol Lett 2019;18:237-46. [PubMed]
- Liu J, Sun J, Zhang Q, et al. shRNA knockdown of DNA helicase ERCC6L expression inhibits human breast cancer growth. Mol Med Rep 2018;18:3490-6. [PubMed]
- Zhang G, Ma J, Xiong J, et al. Upregulation of Excision Repair Cross-Complementation Group 6-Like (ERCC6L) Promotes Tumor Growth in Hepatocellular Carcinoma. Dig Dis Sci 2020. doi: 10.1007/s10620-020-06277-4. [Crossref] [PubMed]
- Yu B, Ding Y, Liao X, et al. Overexpression of PARPBP Correlates with Tumor Progression and Poor Prognosis in Hepatocellular Carcinoma. Dig Dis Sci 2019;64:2878-92. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Goldman M, Craft B, Hastie M, et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv 2019. doi: 10.1101/326470. [Crossref]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507.
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013;41:W169-73. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [Crossref] [PubMed]
- Sticht C, De La Torre C, Parveen A, et al. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 2018;13:e0206239. [Crossref] [PubMed]
- Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 2018;46:D239-45. [Crossref] [PubMed]
- Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 2018;46:D296-302. [Crossref] [PubMed]
- Sauzay C, Petit A, Bourgeois AM, et al. Alpha-foetoprotein (AFP): A multi-purpose marker in hepatocellular carcinoma. Clin Chim Acta 2016;463:39-44. [Crossref] [PubMed]
- Chan AC, Fan ST, Poon RT, et al. Evaluation of the seventh edition of the American Joint Committee on Cancer tumour-node-metastasis (TNM) staging system for patients undergoing curative resection of hepatocellular carcinoma: implications for the development of a refined staging system. HPB (Oxford) 2013;15:439-48.
- Liu PH, Hsu CY, Hsia CY, et al. Prognosis of hepatocellular carcinoma: Assessment of eleven staging systems. J Hepatol 2016;64:601-8. [Crossref] [PubMed]
- Morgan AE, Davies TJ, Mc Auley MT. The role of DNA methylation in ageing and cancer. Proc Nutr Soc 2018;77:412-22. [Crossref] [PubMed]
- O'Neill CP, Dwyer RM. Nanoparticle-Based Delivery of Tumor Suppressor microRNA for Cancer Therapy. Cells 2020;9:521. [Crossref] [PubMed]
- Pandey R, Woo HH, Varghese F, et al. Circulating miRNA Profiling of Women at High Risk for Ovarian Cancer. Transl Oncol 2019;12:714-25. [Crossref] [PubMed]
- Zhang Z, Li J, He T, et al. The competitive endogenous RNA regulatory network reveals potential prognostic biomarkers for overall survival in hepatocellular carcinoma. Cancer Sci 2019;110:2905-23. [Crossref] [PubMed]
- Baumann C, Korner R, Hofmann K, et al. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 2007;128:101-14. [Crossref] [PubMed]
- Leng M, Besusso D, Jung SY, et al. Targeting Plk1 to chromosome arms and regulating chromosome compaction by the PICH ATPase. Cell Cycle 2008;7:1480-9. [Crossref] [PubMed]
- Sridharan V, Azuma Y. SUMO-interacting motifs (SIMs) in Polo-like kinase 1-interacting checkpoint helicase (PICH) ensure proper chromosome segregation during mitosis. Cell Cycle 2016;15:2135-44. [Crossref] [PubMed]
- Kurasawa Y, Yu-Lee LY. PICH and cotargeted Plk1 coordinately maintain prometaphase chromosome arm architecture. Mol Biol Cell 2010;21:1188-99. [Crossref] [PubMed]
- Uehara I, Tanaka N. Role of p53 in the Regulation of the Inflammatory Tumor Microenvironment and Tumor Suppression. Cancers (Basel) 2018;10:219. [Crossref] [PubMed]
- Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ 2019;26:199-212. [Crossref] [PubMed]


