
Integrative bioinformatics analysis of prognostic alternative splicing signatures in gastric cancer
Introduction
Alternative splicing (AS) is a basic and important regulatory mechanism that generates mature mRNAs with different variants in eukaryotic cells (1). These mRNAs are then translated into proteins with similar, different, or even antagonistic functions through RNA splicing (2). AS is involved in the pathogenesis of multiple human diseases, particularly cancers (3). Recent evidence demonstrated that aberrant AS signatures could be a useful target for cancer diagnosis, treatment, and prognosis prediction (4,5).
Gastric cancer (GC) is a common malignant gastrointestinal tumor with poor prognosis, and it is the second leading cause of cancer-related death worldwide (6). Due to its high invasion and metastasis rate, severe clinical symptoms and a low cure rate result in a 5-year survival rate of only ~20%. Remarkably, most of these cases are diagnosed at an advanced stage (7). AS plays a critical role in producing variants associated with gastric carcinogenesis such as CD44 (8), survivin (9), WNT2B (10), and MYH (11). Previous studies mainly focused on alterations at the gene expression level or the differential expressions of AS variants (12). However, studies that aim to identify prognostic biomarkers or therapeutic targets for GC remain limited. Therefore, the prognostic value of AS variants and regulatory splicing factors in gastric carcinogenesis remains elusive. Hence, a comprehensive understanding of AS signatures involved in gastric carcinogenesis will be crucial for prognosis prediction in GC and its effective treatment (13).
Because stomach adenocarcinoma (STAD) is the most prevalent and common pathological type of GC, we profiled genome-wide AS signatures in a cohort of 190 patients with STAD from a large cancer genomic dataset, i.e., the Cancer Genome Atlas (TCGA) project. We aimed to exploit the prognostic impacts of aberrant AS signatures and splicing factors on patients with GC using integrative bioinformatics analysis. The results of this study shed light on the roles of STAD-specific AS signatures, ultimately unraveling their underlying mechanisms in gastric carcinogenesis. We present the following case series in accordance with the STREGA reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-117.
Methods
The research was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval of the study was obtained by TCGA project for this retrospective analysis. It is exempt from consent since it is a retrospective database study.
RNA-seq data acquisition from TCGA
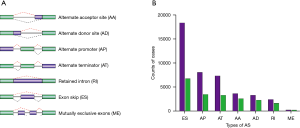
We accessed the TCGA data portal (https://portal.gdc.cancer.gov/) to download STAD RNA-seq data for AS signatures. The SpliceSeq tool, a Java program application, was used to evaluate the types of AS signatures in the STAD cohort. Seven common types of AS signatures have been described: alternate acceptor (AA), exon skip (ES), retained intron (RI), alternate promoter (AP), AA donor (AD), mutually exclusive exon (ME), and alternate terminator (AT). The quantification of these seven common types of AS signatures as a number from 0 to 1 is termed percent-spliced-in (PSI) (14). The demonstrations of these types of AS signatures are shown in Figure 1A.
Survival analysis
The clinical and demographic characteristics of the STAD cohort were selected from the TCGA database. Overall survival (OS) of ≥30 days was used as the inclusion criterion, and 190 patients were enrolled in our research cohort. According to the median cutoff PSI values of each AS signature, these patients were stratified into two groups. We performed univariate Cox regression to analyze OS data and used the results as indicators to weigh the effect of each AS signature on prognosis. Moreover, we performed multivariate Cox regression to determine the independent factors of prognosis and establish prognostic prediction models of the STAD cohort. The survival ROC package (version 1.0.3) in R (version 3.4.3) (15) was used to evaluate the efficiency of every prognostic prediction model by comparing the area under the receiver operator characteristic curve (AUC-ROC) with censored data. Additionally, Kaplan-Meier curves of prognostic predictors were used to predict the 5-year OS of the STAD cohort. Statistical analyses were performed using the Bioconductor package in the R environment (version 3.4.3), and a P value of <0.05 was considered statistically significant (P values were two sided).
Integrative bioinformatics analysis
Instead of a Venn diagram, an UpSet plot was produced by UpSetR (version 1.3.3) to quantitatively analyze >5 interactive sets (16). We used an UpSet plot to present distinguishable relationships between the typical seven types of AS signatures related to OS in this study. After the identification of OS-related genes, they were inputted into the Cytoscape application ReactomeFIViz (17), and a gene network was constructed to search for critical hub genes. Cytoscape (version 3.6.0) was used to visualize splicing correlation networks and elucidate the relationship between PSI values of AS signatures related to GC survival and the expressions of splicing factors. Data analyses were performed with the Bioconductor package in the R environment (version 3.4.3), and a P value of <0.05 was considered statistically significant (P values were two sided).
Results
Integrated AS signatures in the STAD cohort
AS signatures were analyzed in a cohort of 190 patients with STAD obtained from TCGA. Our study identified 43,311 AS signatures in 10,458 genes. We found that ES was the most frequent type of AS signature. In particular, we detected 3,634 AAs in 2,585 genes, 3,292 ADs in 2,261 genes, 8,044 APs in 3,472 genes, 7,335 ATs in 3,286 genes, 18,366 ESs in 6,726 genes, 220 MEs in 209 genes, and 2,420 RIs in 1,618 genes (Figure 1B).
OS-related AS signatures in the STAD cohort
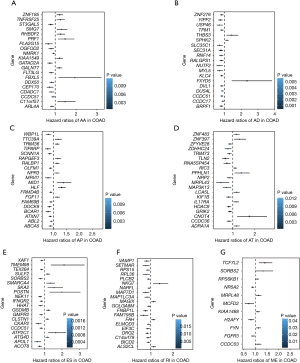
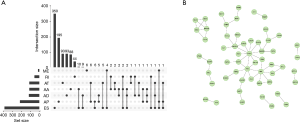
Using univariate Cox analyses to assess the effects of AS signatures on prognosis, we can determine the prognostic value of AS signatures in the STAD cohort. Importantly, a significant correlation was found between 1,308 AS signatures and OS in the STAD cohort (P<0.05). Figure 2 shows the top 20 most critical OS-related AS signatures in the seven types of AS signatures. Notably, most of these AS signatures had a beneficial effect on prognostic factors. It was noticed that one gene could have multiple AS signatures that were simultaneously considerably related to OS. Thus, the UpSet plot presented the intersection of every type of AS signature in the STAD cohort, including overlapping AS signatures (Figure 3A). Intriguingly, most AS-related OS genes are affected by ≥2 types of AS signatures. For instance, AA, AD, and AP in CASP8 were significantly related to OS in the STAD cohort. We next investigated whether OS-related AS genes in the STAD cohort exhibited particular functions. We inputted the most significant OS-related signatures in Reactome to generate gene interaction networks. As shown in Figure 3B, results showed that OS-related genes were associated with network hub genes such as STAT1, FYN, and ABI1.
Prognostic predictors in the STAD cohort
The top 20 meaningful OS-related AS signatures were selected as candidates to detect independent prognostic factors. For determining the seven types of AS signatures in these candidates AS signatures, multivariate analysis based on Cox proportional hazard models was performed to identify independent prognostic factors. Patients of STAD were divided into high- and low-risk groups according to the median value of risk score by the prognostic models. Then, we generated the final multivariate model by combining the candidate AS signatures based on the seven types of AS signatures. Among the seven prognostic prediction models, a single AA model had the most active predictive ability of the analysis results (Figure 4). Remarkably, compared with every type of splicing mode, the analysis results of the STAD cohort showed that the final prognostic predictors have a better predictive effect. Conceivably, the final combined model had an AUC-ROC of 0.948, and the AUC-ROCs of the AA and ES models were 0.914 and 0.841, respectively (Figure 4I). Furthermore, Table 1 provides detailed information on the 17 STAD-specific genes, which are included in the final combinatorial prognostic prediction model.

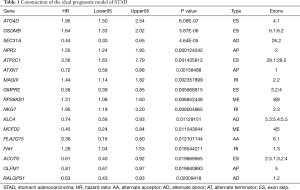
Full table
Potential regulatory network of AS signatures in the STAD cohort
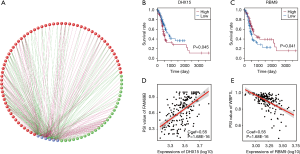
Using gene expression data, we performed a univariate survival test of splicing factors to identify prognosis-related splicing regulators in the STAD cohort. Six splicing factors were detected, which were markedly correlated with OS in the STAD cohort (Figure 5). Moreover, we performed Spearman’s rank correlation to investigate the correlation between these splicing factors and PSI scores of the candidate AS signatures and to build a splicing regulatory network, illustrating significantly correlated relationships (Figure 5A). In the visualization of the correlation networks constructed by Cytoscape, these six OS-related splicing factors (blue dots) were related to 108 OS-related AS signatures, which included 45 adverse signatures (red dots) and 63 favorable AS signatures (green dots). Notably, in most situations, the expression of a splicing factor (grey dot) was considered positively correlated (green lines) with favorable AS signatures (green dots) and negatively correlated (red lines) with adverse AS signatures (red dots). For instance, as revealed in the dot plots, a correlation between AP in FAM69B and DHX15 was observed, suggesting that a high expression of DHX15 is a favorable prognostic factor (Figure 5B,D). Similarly, the correlation between AP in WBP1L and RBM9 was identified, indicating that a high expression of RBM9 might be an adverse prognostic factor (Figure 5C,E).
Discussion
Using TCGA RNA-seq data, we identified the global types of AS signatures in the STAD cohort and obtained systematic insights of their effects on the prognosis of patients with GC (both aberrant AS signatures and splicing factors). Our study identified 1,308 AS signatures in 993 genes that were significantly associated with OS in the STAD cohort. Interestingly, six splicing factors have been suggested to be dysregulated in gastric carcinogenesis, i.e., DHX15, PPP4R2, PRPF38B, RBM9, RBM15, and ILF3. One notable finding was the significant positive correlation between the most favorable prognosis AS signatures and the expression of splicing factors. By contrast, most AS genes associated with poor OS were negatively correlated with the expressions of splicing factors.
Many aberrant genes are known to undergo AS, and these AS variants are implicated in gastric carcinogenesis such as transforming acidic coiled-coil-containing protein 1 (18), human telomerase reverse transcriptase (19), and hepatocyte growth factor (20). The identification of GC-specific AS signatures is essential in ongoing cancer research. However, AD and AA are particularly difficult to detect by traditional microarray analysis because the variable region is often quite small (21). Hence, it is of great importance to highlight the role of high-throughput RNA sequencing to investigate the global alterations in prognostic AS signatures in GC (22). Previous studies comparing patients with primary gastric adenocarcinoma with healthy controls reported alterations in types of AS signatures of approximately 900 genes using TCGA RNA-seq data (23). This transcriptome study revealed exclusive gene features in which AS signatures were misregulated in Epstein-Barr virus-associated GC. Nevertheless, systematic research to identify prognostic AS signatures influencing OS in patients with GC is lacking.
Large clinical samples with different types of AS signatures and AS variant expressions can be obtained from TCGA (24). Thus, this study obtained data from TCGA of a relatively large population to study the role of AS in GC. Our results identified seven typical AS types of the top 20 ranked OS-related AS signatures, most of which were favorable prognostic factors. Related studies have reported that the prognostic value of AS signatures were closely related to the OS of patients with non-small-cell lung cancer and ovarian cancer (25,26). Similar to our study, a previous study reported that the majority of OS-related AS signatures were prognostic factors for both ovarian cancer and lung squamous cell carcinoma (25,26). However, the observation in the current study is inconsistent with earlier findings of lung adenocarcinoma (25).
Similarly, observations of this study confirm the positive association between the most favorable survival prognostic AS genes in GC and the expressions of splicing factors, whereas the majority of poor prognosis and adverse AS signatures are negatively correlated with the expressions of splicing factors. Splicing correlation networks illustrated relationships between the splicing variants of GC-specific genes and splicing factors, adding to the understanding of the possible molecular mechanisms of gastric carcinogenesis.
Previous studies that have demonstrated that alterations in the expressions of splicing factors could lead to the overall changes in the characteristics of AS signatures in various cancers (27). Both of the main families of splicing factors, i.e., serine/arginine-rich proteins (SRPs) and heterogeneous nuclear ribonucleoproteins (hnRNPs), consistently perform opposite functions during RNA splicing (28). SRPs typically interact with splicing-activated elements to promote splice-site recognition, whereas hnRNPs mainly inhibit exon recognition by binding to splicing silencer sequences (29). This study identified three OS-related SRPs (DHX15, PPP4R2, and PRPF38B) and three hnRNPs (RBM9, RBM15, and ILF3), which can potentially be exploited as prognostic biomarkers and therapeutic targets for GC. Correcting these aberrant splicing factors is an imperative issue for future therapeutic approaches.
Moreover, convincing research data obtained in the current study showed that the final prognostic prediction model, which integrates all types of AS signatures, reached an AUC-ROC value of 0.948. Differential types of AS signatures in 17 genes were incorporated into this ideal prognostic prediction model to refine the risk stratification of patients with GC, suggesting useful implications for clinical applications. Indeed, whether these predicted AS isoforms and splicing factors elicit the expected effect needs to be further verified by clinical data, which may not be consistent with in vivo findings. Experimental validation of these potential tumor biomarkers in gastric adenocarcinoma tissues is an imperative issue that needs to be addressed in future studies.
To the best of our knowledge, this study performed the first comprehensive profiling of overall modifications in RNA splicing to identify OS-related AS signatures of GC-specific genes. The results of this study increase our understanding of aberrant AS signatures and splicing regulation, which may ultimately contribute to the development of new treatment strategies for GC.
Conclusions
The study findings contribute to the understanding of aberrant AS signatures and splicing factors in patients with STAD, which can potentially be exploited as prognostic biomarkers and therapeutic targets for GC.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (grant Nos. 81860433), the Natural Science Youth Foundation of Jiangxi Province (Grant Number: 20192BAB215036) and the Foundation for Fostering Young Scholar of Nanchang University (PY201822).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-117
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-117). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval of the study was obtained by TCGA project for this retrospective analysis. It is exempt from consent since it is a retrospective database study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010;463:457-63. [Crossref] [PubMed]
- Jyotsana N, Heuser M. Exploiting differential RNA splicing patterns: a potential new group of therapeutic targets in cancer. Expert Opin Ther Targets 2018;22:107-21. [Crossref] [PubMed]
- Miura K, Fujibuchi W, Sasaki I. Alternative pre-mRNA splicing in digestive tract malignancy. Cancer Sci 2011;102:309-16. [Crossref] [PubMed]
- Le KQ, Prabhakar BS, Hong WJ, et al. Alternative splicing as a biomarker and potential target for drug discovery. Acta Pharmacol Sin 2015;36:1212-8. [Crossref] [PubMed]
- Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene 2015;34:1-14. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol 2016;22:2403-14. [Crossref] [PubMed]
- da Cunha CB, Oliveira C, Wen X, et al. De novo expression of CD44 variants in sporadic and hereditary gastric cancer. Lab Invest 2010;90:1604-14. [Crossref] [PubMed]
- Krieg A, Mahotka C, Krieg T, et al. Expression of different survivin variants in gastric carcinomas: first clues to a role of survivin-2B in tumour progression. Br J Cancer 2002;86:737-43. [Crossref] [PubMed]
- Katoh M. Differential regulation of WNT2 and WNT2B expression in human cancer. Int J Mol Med 2001;8:657-60. [PubMed]
- Tao H, Shinmura K, Hanaoka T, et al. A novel splice-site variant of the base excision repair gene MYH is associated with production of an aberrant mRNA transcript encoding a truncated MYH protein not localized in the nucleus. Carcinogenesis 2004;25:1859-66. [Crossref] [PubMed]
- Tang X, Li J, Yu B, et al. Osteopontin splice variants differentially exert clinicopathological features and biological functions in gastric cancer. Int J Biol Sci 2013;9:55-66. [Crossref] [PubMed]
- Li Y, Yuan Y. Alternative RNA splicing and gastric cancer. Mutat Res 2017;773:263-73. [Crossref] [PubMed]
- Ryan MC, Cleland J, Kim R, et al. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics 2012;28:2385-7. [Crossref] [PubMed]
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000;56:337-44. [Crossref] [PubMed]
- Lex A, Gehlenborg N, Strobelt H, et al. UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 2014;20:1983-92. [Crossref] [PubMed]
- Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol 2010;11:R53. [Crossref] [PubMed]
- Line A, Slucka Z, Stengrevics A, et al. Altered splicing pattern of TACC1 mRNA in gastric cancer. Cancer Genet Cytogenet 2002;139:78-83. [Crossref] [PubMed]
- Xu JH, Wang YC, Geng X, et al. Changes of alternative splicing variants of human telomerase reverse transcriptase during gastric carcinogenesis. Ai Zheng 2008;27:1271-6. [PubMed]
- Smyth EC, Sclafani F, Cunningham D. Emerging molecular targets in oncology: clinical potential of MET/hepatocyte growth-factor inhibitors. Onco Targets Ther 2014;7:1001-14. [Crossref] [PubMed]
- Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature 2008;456:470-6. [Crossref] [PubMed]
- Feng H, Qin Z, Zhang X. Opportunities and methods for studying alternative splicing in cancer with RNA-Seq. Cancer Lett 2013;340:179-91. [Crossref] [PubMed]
- Armero VES, Tremblay MP, Allaire A, et al. Transcriptome-wide analysis of alternative RNA splicing events in Epstein-Barr virus-associated gastric carcinomas. PLoS One 2017;12:e0176880. [Crossref] [PubMed]
- Casamassimi A, Federico A, Rienzo M, et al. Transcriptome Profiling in Human Diseases: New Advances and Perspectives. Int J Mol Sci 2017.18. [PubMed]
- Li Y, Sun N, Lu Z, et al. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett 2017;393:40-51. [Crossref] [PubMed]
- Zhu J, Chen Z, Yong L. Systematic profiling of alternative splicing signature reveals prognostic predictor for ovarian cancer. Gynecol Oncol 2018;148:368-74. [Crossref] [PubMed]
- Dvinge H, Kim E, Abdel-Wahab O, et al. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer 2016;16:413-30. [Crossref] [PubMed]
- Kedzierska H, Piekielko-Witkowska A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett 2017;396:53-65. [Crossref] [PubMed]
- Bonomi S, Gallo S, Catillo M, et al. Oncogenic alternative splicing switches: role in cancer progression and prospects for therapy. Int J Cell Biol 2013;2013:962038. [Crossref] [PubMed]


