Down-regulation of small nuclear RNA (snRNA) RNU5E-1 in hepatocellular carcinoma presents with vital clinical significance
Introduction
Over the past 20 years, hepatocellular carcinoma (HCC), the most common form of liver cancer, has become the fourth causing of cancers-related death worldwide and the fifth diseases-related mortality in China (1,2). With the development and advancement of rational approaches to prevention, surveillance, early detection, diagnosis and treatment, some efficacy has been showed on HCC overall mortality (2). However, HCC incidence and cancer-specific mortality has being still increasing in many countries, and the most of HCC patients has been diagnosed at an advanced stage as before in many parts of the world (3). In recent clinical practice, many diagnostic and prognostic bio-markers (serum or tissue), such as alpha fetoprotein (AFP) and carcinoembryonic antigen (CEA), has been found and used in liver disease wildly but they perform with low sensitivity and specificity (4). Therefore, appropriately accurate and prognosis biomarkers for HCC are urgently needed, and it is of great importance to identify some reliable early-stage biomarkers for accurate HCC diagnosis and prediction of patient’s long-term outcome so as to reduce the cancers-related death burden (4,5).
Small nuclear RNAs (snRNAs), belonging to non-coding RNAs (ncRNAs), comprise a cluster of highly abundant transcripts in nucleoplasm. Generally, these snRNAs, implicated in vital regulation of post-transcriptional process, are recognized to assemble with associated proteins into spliceosomes and then make pre-mRNAs functional (6). Sometimes, dysregulation of some specific snRNAs can make wrong in biological process of ribonucleoprotein biogenesis, mRNA maturation and protein synthesis. And abnormal protein metabolism will lead to various diseases, including tumorigenesis, through diverse mechanisms (7). In recent years, emerging evidence has proved that snRNAs can perform vital roles in the development as well as clinical assessment of malignant tumors. For instance, the dysfunction of H/ACA small nucleolar RNAs (snoRNAs) induced by oncogenic RAS gene inhibited two pseudouridine modifications, and promoted HCC development by increasing the translational miscoding in HCC cells (8). Besides, Peng et al. demonstrated that snRNA SNHG16 could aggravate the epithelial-mesenchymal transition of bladder cancer through miR-17-5p/TIMP3 signaling (9). Moreover, it has been proved that the upregulation of circulating snRNA RNU2-1f could function as a diagnostic and prognostic biomarker for lung cancer, pancreatic and colorectal adenocarcinoma patients (10,11). Therefore, many snoRNAs, which possess potential correlation with HCC and other malignant tumors, are needed to be revealed.
The snRNA U5E-1 (RNU5E-1), a new variant of U5 snRNA, was firstly identified and characterized from HeLa cells (12). It will be assembled into small nuclear ribonucleoprotein (snRNP) complex to splice heterogeneous nuclear RNAs (hnRNA), which are precursors of the mRNAs. Thus, abnormality of RNU5E-1 will possibly disturb the process of DNA transcription and lead to cellular dysfunction. However, there remains no researches to explore the potential relationship between RNU5E-1 expression and any malignant diseases.
In this study, we aimed to explore the relative expression of RNU5E-1 in HCC cells and tissues. Then, we explored the accurate diagnostic and prognostic predicting abilities of RNU5E-1 for HCC patients.
We present the following article in accordance with the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-49.
Methods
Patient data and specimens
In total, 100 inpatients undergoing hepatectomy at the First Affiliated Hospital, School of Medicine, Zhejiang University from January 2013 to August 2014 were included into the present study. With use of hospital medical database system, the perioperative data of patients were collected, including age, gender, hepatitis B virus infection, liver cirrhosis, AFP, tumor diameter, TNM stages and so on. After hepatectomies, the liver lesions of patients were sent to the histological examinations, and the final reports were reviewed by the identical experienced pathology doctor.
All patients were diagnosed with HCC according to the histological examination of tumor lesions. The HCC specimens and corresponding adjacent tissues of them were obtained from surgical resections and all isolated samples preserved in liquid nitrogen were transported to laboratory within 30 min. Valid survival data was obtained over a period of 60 months.
Cell culture
The human normal hepatocyte cell line QSG-7701 and several human HCC cell lines (SK-HEP-1, Hep 3B and Huh-7) were cultured in DMEM (Gibco, USA) containing heat-inactivated 10% fetal bovine serum (FBS, Gibco, USA), 100 mg/mL streptomycin and 100 U/mL penicillin in a humidified atmosphere of 5% CO2 at 37 °C.
RNA extraction and cDNA synthesis
Total RNA was extracted from cells, tumor and adjacent tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The concentration of RNA was measured by Nanodrop 2000 spectrophotometer (Thermo Scientific Inc., Waltham, MA, USA). The RNA was reversely transcribed into cDNA using HiScript II Reverse Transcriptase SuperMix with gDNA wiper (Vazyme, Nanjing, China) according to the manufacturer’s instructions.
Real-time PCR analysis
Quantitative real-time polymerase chain reaction (RT-PCR) was implemented to determine the expression level of RNU5E-1 using Fast Start Universal SYBR Green Master ROX (Roche, Basel, Switzerland) following the manufacturer’s instructions. The sequences of all used PCR primers were as follows (5'-3'): RNU5E-1: CCGTGGAGAGAAACGAGTGTG (forward), CCTGCTAAAAAACGCAAGGC (reverse); GAPDH: CAGGAGGCATTGCTGATGAT (forward), GAAGGCTGGGGCTCATTT (reverse). Combined with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression as the endogenous control, relative RNU5E-1 expression was calculated by the comparative Ct method formula 2−∆∆Ct. Every sample was tested in triplicates.
Statistical analysis
Generally, the continuous variables were expressed as mean value with standard deviation or number (percentage). Statistical software package SPSS19.0 (SPSS Inc., Chicago, IL, USA) was used to accomplish all statistical analysis. Quantitative variables were assessed by Student’s t-test, while Chi-square test and Fisher’s exact test were applied to analyze qualitative and categorical data. Overall survival curves were plotted with use of the Kaplan-Meier method. Overall, the association of tumor-free survival time and long-term survival was plotted with use of Cox univariate analysis and multivariate analysis. All statistical differences of P<0.05 were considered statistically significant.
The study was in accordance with the ethical guidelines of the 2013 Declaration of Helsinki. Ethical approval was obtained from the Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University (NO.: 20192). All data were analyzed anonymously and identified prior to analysis. Written informed consent was obtained from the patient for publication of this study and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Results
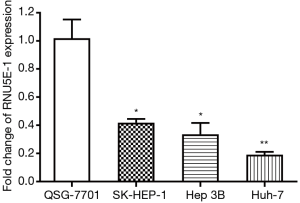
The down-regulation of RNU5E-1 in HCC cell lines
To investigate RNU5E-1 levels in HCC, we performed RT-PCR analysis for the expression of RNU5E-1 in human normal hepatocyte cell line QSG-7701 as well as various HCC cell lines (SK-HEP-1, Hep 3B and Huh-7). According to the result in Figure 1, RNU5E-1 was sharply down-expressed in several HCC cell lines compared to normal liver cell line. Recognizing QSG-7701 as control, the fold change values of RNU5E-1 expression in HCC cells were 0.417 (SK-HEP-1, P<0.05), 0.313 (Hep 3B, P<0.05) and 0.189 (Huh-7, P<0.01), respectively.
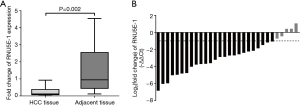
The down-regulation of RNU5E-1 in HCC tissues
To further definite the association between RNU5E-1 expression and HCC, we calculated RNU5E-1 expression in paired HCC lesions and corresponding adjacent liver tissues. As showed in Figure 2A, RNU5E-1 was obviously down-regulated in HCC tissues compared to matched adjacent liver tissues (average fold change =0.322, P=0.002). Among that, RNU5E-1 was down-expressed by at least twofold in 83.3% paired HCC tissues (Figure 2B). In accordance with the verification results of both cell lines and tissues, it implied that RNU5E-1 might function as a tumor suppressor in the progress of HCC.
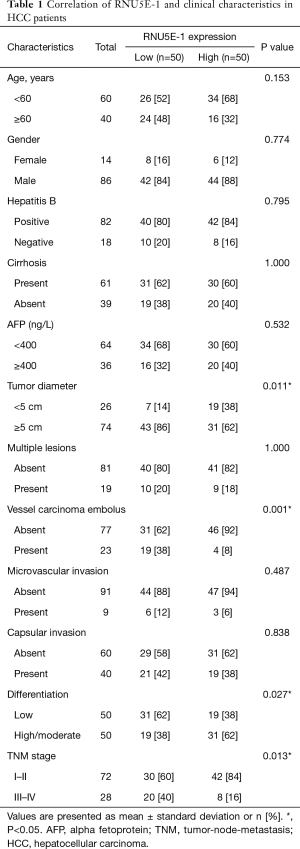
Correlation of RNU5E-1 and clinical characteristics in HCC patients
To gain further insights into the correlation of RNU5E-1 expression and HCC clinical characteristics, patients were finally sorted into high or low expression groups and clinico-pathological data was summarized in Table 1. By comparing the patients in high and low RNU5E-1 expression groups, we found that patients in low RNU5E-1 expression group exhibited significantly worse tumor characteristics, including larger tumor lesions (tumor diameter >5 cm, P=0.011) and more vessel carcinoma embolus (P=0.001). Besides, HCC patients with down-regulated RNU5E-1 expression had more severe status in classification of TNM stages (P=0.013) as well as differentiation degree of HCC (P=0.027). However, there was no significant difference in aspects of age, gender, hepatitis B infection, liver cirrhosis, AFP and so on. These data convincingly revealed that RNU5E-1 expression was remarkably related to occurrence and development of HCC.
Full table
Correlation of RNU5E-1 expression level and disease-free survival in HCC patients
In order to evaluate the potential ability of RNU5E-1 in predicting patient’s tumor recurrence, Kaplan-Meier analysis was also performed in aspects of postoperative disease-free survival time. Notably, HCC patients with lower RNU5E-1 expression obviously showed higher tumor recurrence rate and shorter disease-free survival time (average disease-free survival time: low vs high RNU5E-1 group, 16.6 vs. 43.9 months, P<0.001, Figure 3). In the following univariate and multivariate Cox regression analysis, we found that RNU5E-1 expression, TNM stage, pathological differentiation, vessel carcinoma embolus as well as tumor diameter all presented with independent capability in predicting disease-free survival of HCC patients (P<0.05, Table 2).
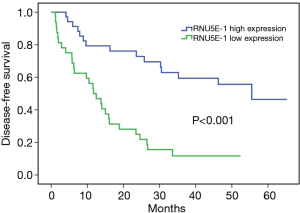
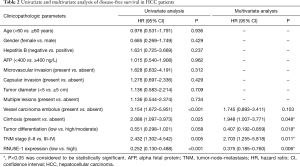
Full table
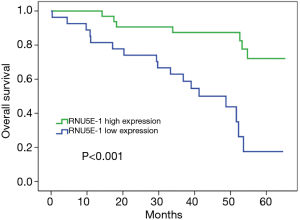
Correlation of RNU5E-1 expression level and overall survival in HCC patients
Moreover, there was effectively demonstrated that HCC patients with low RNU5E-1 expression level were inclined to get poorer overall survival time during 5 years of postoperative follow-up (average overall survival time: low vs high RNU5E-1 group, 39.3 vs. 57.9 months, P<0.001, Figure 4). Similarly, through univariate and multivariate Cox regression analysis, we certified that RNU5E-1 expression could be used as an independent prognostic indicator to assess the overall survival in HCC patients (P<0.05, Table 3).
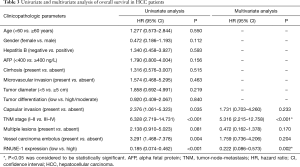
Full table
Discussion
For lack of accurate early screening methods, HCC has become one of great burdens with high morbidity and poor long-term survival in the world (13-15). Although a great number of biomarkers have been explored for early diagnosis and long-term prognosis of HCC, ideal indicator with high sensitivity and specificity remains blank (16,17). Thus, more novel and effective gene biomarkers need to be explored so as to benefit HCC patients.
With the development of advancing high-throughput gene sequencing techniques, plenty of snRNAs have been identified. Commonly, snRNAs are recognized to assemble with associated proteins into spliceosomes in charge of pre-mRNAs splicing. Among that, however, the spliceosomal uridylic acid rich snRNAs, including U1, U2, U4, U5 and U6, have been proved directly correlating with development of malignant tumors (18). In the research of Mukherjee et al., they found that an overall significant quantitative alteration of major snRNAs U4, U5, and U6 was identified and it contributed to the process of neoplastic transformation (19). Besides, according to the study of Hamada, a specific partial sequence of U5 snRNA, designated as RNA3S, was found to confer growth advantages and promote epithelial-mesenchymal transition in human HeLa cells (20). And it was also elucidated that altered expression in a transforming sequence of U5 snRNA could inhibit gap junction during carcinogenesis (21,22). What’s more, U2-1f snRNA could function as a diagnostic and prognostic biomarker for lung cancer, pancreatic as well as colorectal adenocarcinoma (10,11). Considering these significant relationships between uridylic acid rich snRNAs and malignant tumors, some snRNAs might be explored as potential biomarkers for HCC clinical management.
According to gene database, RNU5E-1, one newly identified variant of U5 snRNAs, locates in chromosomal locus 1p36. Although there is no evidence revealing the biological role of RNU5E-1 in tumorigenesis, several studies indicated that chromosomal locus 1p36 is closely associated with HCC characteristics. Recently, Zhang et al. reported that chromosomal locus 1p36 was a new locus attaching great importance to HCC in chronic hepatitis B virus carriers, which directly correlated with several tumorigenesis pathways like KIF1B, UBE4B and PGD (23). In addition, snoRNA runt-related transcription factor 3 gene (RUNX3), derived from chromosomal locus 1p36, was found to dysfunction in HCC lesions and showed as a tumor suppressor gene related to hepatocarcinogenesis (24). Therefore, we suspected that dysregulation of snoRNA from chromosomal locus 1p36 might be strongly associated with HCC occurrence and development as negative regulatory factor, which was consistent with our findings about the down-regulation of RNU5E-1 in HCC. Besides, RNU5E-1 is identified as one variant of U5 snRNAs, which are assembled into the snRNP complex to splice the precursors of mRNAs (12). Thus, abnormal RNU5E-1 regulation will possibly disturb cellular mRNA maturation and transcription process, and finally leads to malignant tumorigenesis. However, to validate the exact action mechanisms of RNU5E-1 in HCC, more researches are needed in the future.
In conclusion, we for the first time explored the relationship between RNU5E-1 expression and HCC in this study. And we found that RNU5E-1 was significantly down-regulated in HCC cell lines as well as HCC tissues. Moreover, RNU5E-1 expression was significantly related to tumor size, vessel carcinoma embolus, differentiation level and TNM stages. As for the follow-up of HCC patients, low RNU5E-1 expression was independently associated with both severe tumor-free and long-term survival. Therefore, RNU5E-1 could function as a potential indicator for diagnosis as well as prognostic prediction of HCC.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (No. 81572307, 81773096), Major Project of Medical and Health Technology Development Program in Zhejiang Province (No. 7211902), Key Research and Development Project of Zhejiang Province (No. 2018C03085), Public Welfare Technology Research Project of Zhejiang Province (No. LGD19C040006) and General Research Project of the Zhejiang Provincial Education Department (No. Y201840044).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-49
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-49). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was in accordance with the ethical guidelines of the 2013 Declaration of Helsinki. All participants had signed written informed consent. Ethical approval was obtained from the Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University (No.: 20192). All data were analyzed anonymously and identified prior to analysis.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhou M, Wang H, Zeng X, et al. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019;394:1145-58. [Crossref] [PubMed]
- Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589-604. [Crossref] [PubMed]
- Fitzmaurice C, Allen C, Barber RM, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2017;3:524-48. [Crossref] [PubMed]
- Berretta M, Cavaliere C, Alessandrini L, et al. Serum and tissue markers in hepatocellular carcinoma and cholangiocarcinoma: clinical and prognostic implications. Oncotarget 2017;8:14192-220. [Crossref] [PubMed]
- Long J, Zhang L, Wan X, et al. A four-gene-based prognostic model predicts overall survival in patients with hepatocellular carcinoma. J Cell Mol Med 2018;22:5928-38. [Crossref] [PubMed]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 2007;8:209-20. [Crossref] [PubMed]
- Carey KT, Wickramasinghe VO. Regulatory Potential of the RNA Processing Machinery: Implications for Human Disease. Trends Genet 2018;34:279-90. [Crossref] [PubMed]
- McMahon M, Contreras A, Holm M, et al. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. Elife 2019;8:e48847. [Crossref] [PubMed]
- Peng H, Li H. The encouraging role of long noncoding RNA small nuclear RNA host gene 16 in epithelial-mesenchymal transition of bladder cancer via directly acting on miR-17-5p/metalloproteinases 3 axis. Mol Carcinog. 2019;58:1465-80. [Crossref] [PubMed]
- Köhler J, Schuler M, Gauler TC, et al. Circulating U2 small nuclear RNA fragments as a diagnostic and prognostic biomarker in lung cancer patients. J Cancer Res Clin Oncol 2016;142:795-805. [Crossref] [PubMed]
- Baraniskin A, Nöpel-Dünnebacke S, Ahrens M, et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int J Cancer 2013;132:E48-57. [Crossref] [PubMed]
- Sontheimer EJ, Steitz JA. Three Novel Functional Variants of Human U5 Small Nuclear RNA. Mol Cell Biol 1992;12:734-46. [Crossref] [PubMed]
- Sayiner M, Golabi P, Younossi ZM. Disease Burden of Hepatocellular Carcinoma: A Global Perspective. Dig Dis Sci 2019;64:910-7. [Crossref] [PubMed]
- Mak LY, Cruz-Ramón V, Chinchilla-López P, et al. Global Epidemiology, Prevention, and Management of Hepatocellular Carcinoma. Am Soc Clin Oncol Educ Book 2018;38:262-79. [Crossref] [PubMed]
- Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019;156:477-491.e1. [Crossref] [PubMed]
- Song P, Tang Q, Feng X, et al. Biomarkers: evaluation of clinical utility in surveillance and early diagnosis for hepatocellular carcinoma. Scand J Clin Lab Invest Suppl 2016;245:S70-6. [Crossref] [PubMed]
- Tsuchiya N, Sawada Y, Endo I, et al. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol 2015;21:10573-83. [Crossref] [PubMed]
- Dvinge H, Guenthoer J, Porter PL, et al. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res 2019;29:1591-604. [Crossref] [PubMed]
- Mukherjee S, Manna S, Mukherjee P, et al. Differential alterations in metabolic pattern of the spliceosomal uridylic acid-rich small nuclear RNAs (UsnRNAs) during malignant transformation of 20-methylcholanthrene-induced mouse CNCI-PM-20 embryonic fibroblasts. Mol Carcinog 2009;48:773-8. [Crossref] [PubMed]
- Hamada K. Morphological transformation caused by a partial sequence of U5 small nuclear RNA. Mol Carcinog 1997;20:175-88. [Crossref] [PubMed]
- Hayashi T, Trosko JE, Hamada K. Inhibition of gap junctional intercellular communication in rat liver epithelial cells with transforming RNA. FEBS Lett 2001;491:200-6. [Crossref] [PubMed]
- Hamada K, Kumazaki T, Satoh S. Effect of transforming RNA on the synthesis of a protein with a secretory signal sequence in vitro. J Biol Chem 1999;274:15786-96. [Crossref] [PubMed]
- Zhang H, Zhai Y, Hu Z, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet 2010;42:755-8. [Crossref] [PubMed]
- Mori T, Nomoto S, Koshikawa K, et al. Decreased expression and frequent allelic inactivation of the RUNX3 gene at 1p36 in human hepatocellular carcinoma. Liver Int 2005;25:380-8. [Crossref] [PubMed]