Could CTSK and COL4A2 be specific biomarkers of poor prognosis for patients with gastric cancer in Asia?—a microarray analysis based on regional population
Introduction
Nowadays, it is widely considered that gastric cancer (GC) is an inflammation-driven disease, Helicobacter pylori and Epstein-Barr (EB) virus infections are thought to be the important risk factors of GC. Chronic inflammation in gastric mucosa leading to changes in microenvironment followed by molecular alterations, causes neoplasia gradually, but the specific mechanisms are still uncertain (1). The proposition of molecular subtypes of GC not only analyzes the molecular changes of GC and its corresponding biological behavior characteristics from genetic level, but also provides a favorable guidance for the selection of anti-tumor drugs (2-4). Other genes used for grouping GC have been focused by several studies in order to guide GC treatment and evaluate prognosis as well, such as human epidermal growth factor receptor 2 (HER2) and tumor protein 53 (TP53) (5,6). But beyond that, ethnic differences in response to anti-tumor therapies in GC patients are always concerned. For instance, patients from Asia have a better prognosis and response to treatments than Caucasian (7,8); survival differences are independent of clinical and pathological factors among different races and ethnicities patients (9); and significant differences found in the frequencies of somatic mutation from diverse geographic populations (10). Accordingly, GC is highly heterogeneous, and its biological behavior has wide genetic and epigenetic differences between different individuals or between different lesions of the same individual, which might further result in different prognosis and treatment outcome. Therefore, it is quite necessary to explore the genetic changes and potential molecular mechanisms of the occurrence and development of GC from the aspects of race and region. On one hand, it can help us to find more specific biomarkers for diagnosis and assessing prognosis. On the other hand, it has a very significant guiding role for us to better design individualized regimens, especially targeting therapies (11).
To find reliable biomarkers, it is a feasible method to obtain gene expression profiles of GC from public functional genomics databases like GEO (12) and TCGA, and then perform bioinformatics analysis. In this study, we selected GC expression profiles of Asian population in GEO database, screened out hub genes for GC patients through microarray analysis. For one, the work helps us to understand the genetic changes clearly in GC groups in Asia. For another, during the analysis, we mainly discuss whether COL4A2 and CTSK can be used as effective biomarkers for early diagnosis, prognosis assessment, recurrence monitoring, and therapeutic target in patients with GC in Asia. And the work offers a new direction for the diagnosis and treatment of GC.
Methods
Microarray data
Datasets were retrieved from GEO database (https://www.ncbi.nlm.nih.gov/geo/) with “gastric cancer” as the key word. The filters of organism and study type were limited as “Homo sapiens” and “Expression profiling by array”, respectively. Then the gene expression matrix and the corresponding platform TXT files of target datasets were downloaded. R software (R 3.6.1, https://cran.r-project.org/) and related packages (http://www.bioconductor.org/) were used for data processing. The datasets utilized to differential analysis must be satisfied the following conditions: (I) all of the sequencing samples were from the patients with GC in China; (II) the datasets for differential gene analysis contained controls for cancer and cancer-adjacent tissues; (III) the sample size of each dataset was at least 20; (IV) the information of the platform annotation was available. The datasets used for prognosis analysis must be contained detailed survival data.
Screening for DEGs
A volcano map was plotted to assess the differential expression of all genes by the ggplot2 package (https://cran.r-project.org/web/packages/ggplot2/index.html). The limma package was used for screening the DEGs in candidate dataset (13). As a DEG, it is necessary to satisfy both statistical P value <0.05 and |log fold change (FC)| >1. The co-expressed genes of DEGs were visualized by UpSetR package (14).
Construction of PPI network and molecular modules analysis
The PPI network of DEGs was constructed in the STRING database (https://string-db.org/) through the following setting: meaning of network edges was set as “confidence”, minimum required interaction score was selected as “medium confidence (0.400)” and finally display simplification was to hide disconnected nodes in the network (15). The PPI-data was downloaded and then identified hub genes and molecular modules by using the cytoHubba and MCODE plug-in Cytoscape software (version 3.7.1, https://cytoscape.org/), respectively. In MCODE, filters were based on the default parameters as “Degree Cutoff =2,” “Node Score Cutoff =0.2,” “K-Core =2” and “Max.Depth =100” (16).
GO and KEGG pathway analyses
Functional annotation and pathway analyses for DEGs were done in the Enrichr database (http://amp.pharm.mssm.edu/Enrichr/), which is a comprehensive database for GO and KEGG pathway enrichment analysis (17). And the results of which were visualized by using GOplot package and Cytoscape software, respectively. Adjusted P value <0.05 was as a selecting criterion.
Prognosis analyses
Firstly, survival analysis was conducted for DEGs using the Kaplan Meier analysis in the one of the datasets from GEO database so as to identify hub genes associated with prognosis of GC. Secondly, the racial survival differences of hub genes were evaluated again in the Kaplan-Meier plotter database (http://kmplot.com/analysis/), which included gene chip and RNA-seq data-sources from GEO database and TCGA (18). Thirdly, the hub genes were re-analyzed the prognostic value in multiple datasets from different cohort in GEO database. The results were visualized by forestplot (https://cran.r-project.org/web/packages/forestplot) and survival package (https://cran.r-project.org/web/packages/survival/) in R.
TIMER database analysis
The correlations between the hub genes expression and the infiltration levels of inflammatory cells, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, were analyzed based on TIMER database (19). Moreover, we also explored the relationship of hub genes expression and molecular markers that had been reported in published studies (20,21), including markers of tumor-associated macrophages (TAMs), M2 macrophages, myeloid-derived suppressor cells (MDSCs), natural killer (NK) cells, dendritic cells (DCs), regulatory T cells (Tregs), and exhausted T cells. Correlation strength was classified according to the absolute value of partial correlation coefficient as follow: 0.00–0.19 “very weak”, 0.20–0.39 “weak”, 0.40–0.59 “moderate”, 0.60–0.79 “strong”, 0.80–1.0 “very strong” (22). The gene expression level was displayed with log2 RSEM.
Results
Microarray data
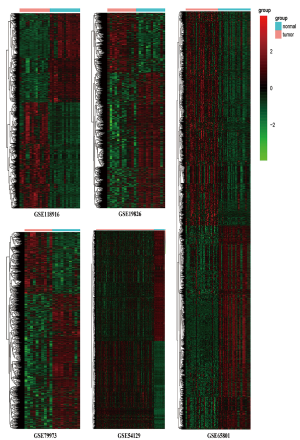
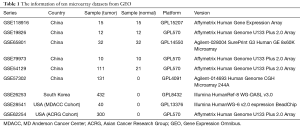
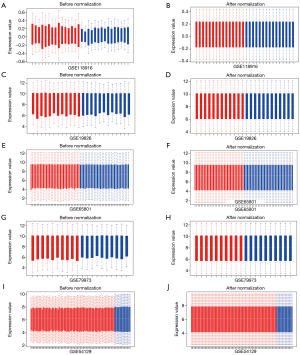
The GSE118916 (23), GSE19826 (24), GSE65801 (25), GSE79973 (26,27), and GSE54129 dataset were used for differential analysis, including 90 normal gastric tissue samples and 180 GC samples of China. The GSE57302 (28), GSE26253 (29,30), GSE28541 (29) and GSE62254 (6,29) dataset were used for prognosis analysis. The information of ten datasets was shown in Table 1. The five datasets for differential gene analysis were normalized, which of the results were revealed in Figure 1.
Full table
Identification of DEGs
The DEGs from five dataset was shown in Figure 2A,B,C,D,E. Two hundred twenty-two co-expressed genes were obtained by integrating bioinformatics analysis of all DEGs, covering 82 up-regulated expression genes and 140 down-regulated expression genes. The number of all and co-expressed DEGs were shown in Figure 2F. The co-expressed genes were provided in Table S1. The cluster heat map of all genes from five datasets was shown in Figure S1.
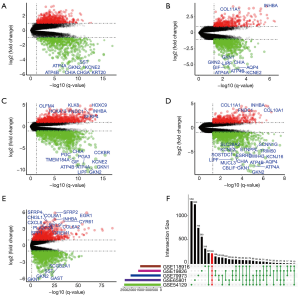
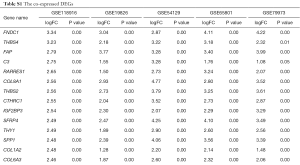
Full table
PPI network and molecular module
The PPI network of 222 integrated DEGs was built through STRING database and the result was shown in Figures 3A,S2. The top 20 hub genes were FN1, COL3A1, COL1A2, COL5A2, BGN, FBN1, THBS2, TIMP1, SPARC, COL6A3, COL5A1, SPP1, CDH11, COL12A1, VCAN, PDGFRB, COL4A2, ASPN, SERPINH1, COL10A1 and the matching information from GSE118916 was listed in Table 2. Six molecular modules were identified by using MCODE, the most important of which contained 32 genes, as visualized in Figure 3B.
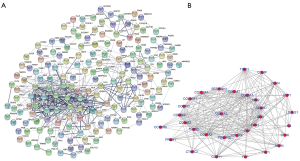

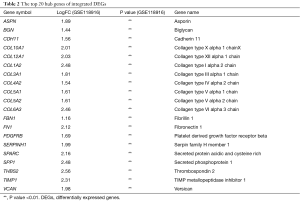
Full table
Functional enrichment analyses of DEGs
The total results of GO functional analyses for DEGs were shown in Table S2. It was evidence that the top 20 hub genes were mainly involved in the biological process (BP) of cell metabolism and the formation of extracellular matrix (ECM). Simultaneously, the molecular functions of hub genes were enriched in various binding of BP, as shown in Figure 4. Beyond that, the KEGG pathway enrichment of DEGs were mainly focus on protein digestion and absorption, ECM-receptor interaction, focal adhesion (FA) and phosphoinositide 3-kinase (PI3K)-protein kinase B (AKT) signaling pathway, as shown in Figure 5 and Table S3.

Full table
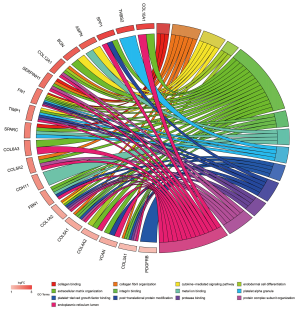
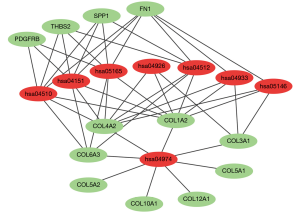
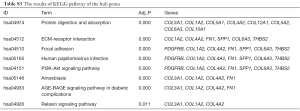
Full table
Prognosis analysis of DEGs
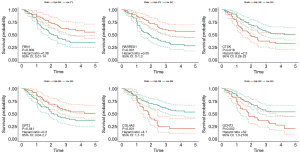
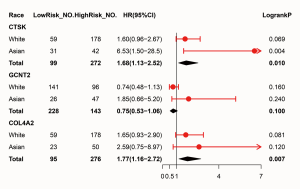
It turned out that six genes were associated with prognosis of GC patients from GSE57302, and of which FBN1 (P=0.009), RARRES1 (P=0.001), GPT2 (P=0.041) were related to good prognosis in GC patients, while COL4A2 (P<0.001), CTSK (P=0.018), GCNT2 (P=0.002) were related to poor prognosis (Figure 6). To further examine the prognostic potential of CTSK, COL4A2 and GCNT2 in different races of GC, we assessed the correlation of the OS and these genes expression between Asian and White patients in the Kaplan-Meier plotter databases, and found that over-expressions of CTSK and COL4A2 reflected a worse OS in total patients (P<0.05). Specifically, it was worth mentioning that the CTSK expression level was correlated with worse OS in Asian GC patients (HR =6.53, P=0.01), but was not associated with OS of White patients (HR =1.60, P=0.069) (Figure 7). Then we continued to re-analyze the survival differences of CTSK and COL4A2 in the GSE26253, GSE28541 and GSE62254 dataset and the eventual outcomes also showed that the high-expression of CTSK and COL4A2 had a significant association with poor prognosis in the cohort of Asian GC patients (Figure 8).
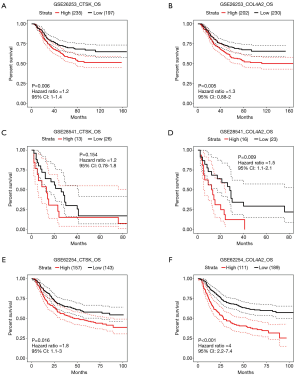
The relationship between CTSK, COL4A2 and GCNT2 expression level and inflammatory cell infiltration
We analyzed whether the expression of CTSK, COL4A2 and GCNT2 were correlated with inflammatory cell infiltration levels in GC. The results showed that the level of CTSK expression had significant correlations with tumor purity, macrophage, neutrophil and dendritic cell (Figure 9). We also investigated the relationships between CTSK expression and biomarkers of different immune cells, included TAMs, M2 macrophages, MDSCs, NK cells, DCs, Tregs and exhausted T cells. We found that CCL2, IL10 of TAMs, CD163, VSIG4, MS4A4A of M2 phenotype, CD33, ITGAM, CD14, CSF1R of MDSCs, NRP1, IL3RA, ITGAX of DCs, TGFB1 of Tregs, and TIM3 of T cell exhaustion were significantly correlated with CTSK expression in GC (P<0.0001; Table 3).
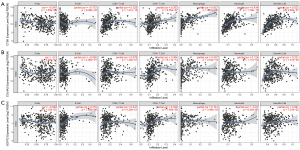
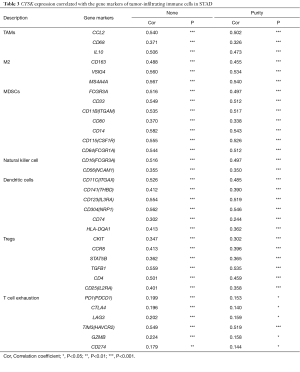
Full table
Discussion
The occurrence of tumors shows significant racial differences (31). Changes in genetic level and ethnic heterogeneities have always been the focus of tumor researches. To improve early diagnostic rate, and screen for therapeutic targets on ethnic characteristics for GC, it is urgently necessary to identify sensitive and specific prognostic biomarkers.
In our study, with the differential analysis of five datasets comprising the GSE118916, GSE19826, GSE65801 GSE79973 and GSE54129 by using limma package in R software, and 222 co-expressed DEGs were found. Then, a functional analysis, a PPI network and the most important molecular module of DEGs were performed through online databases and Cytoscape software. A prognostic analysis of DEGs was conducted by using survival package in R software, which of the results showed that CTSK (Cathepsin K, CatK), GCNT2 [Glucosaminyl (N-acetyl) transferase2] and COL4A2 (Collagen type IV alpha 2 chain) were related to poor prognosis, while FBN1 (Fibrillin 1), RARRES1 (Retinoic acid receptor responder 1), GTP2 (Glutamic pyruvate transaminase 2) were associated with desirable prognosis in GC patients in Chinese population. Furthermore, we found that the high-expression levels of CTSK and COL4A2 are significantly correlated with poor prognosis of GC in Asian through cross-validation between databases.
CTSK, belonging to the cathepsin L-like cluster of C1A family, has been confirmed to play a key role in ECM-remodeling, regulation of cytokine level and tumor growth factor, process of lymph node and bone metastasis in a variety of cancers such as breast and prostate cancer (32-35). In gastric and oral squamous cell carcinoma, researchers also report that up-regulation of CTSK associates with lymph node metastasis and poor prognosis (36,37). In terms of therapeutic significance, CTSK may be a therapeutic target for patients with prostate cancer at risk of bone metastases (33). Alongside these function, we also found that CTSK has significant correlations not only with macrophages, neutrophils and DCs in TME, but also with numerous gene marker sets of various inflammatory cells in GC, for instance CCL2, IL10, VSIG4, MS4A4A, CD33, ITGAM, CD14, CSF1R, NRP1, IL3RA, ITGAX, TGFB1, CD4, and TIM3. Although T cells recruited to TME have potential to kill tumor cells, more often, they are powerless, fading and exhausting due to high expression of genes like TIM3 (38). TAMs, particularly M2 phenotype, play a vital role in promoting tumorigenesis through inducing neovascularization, regulating inflammatory responses and the reconstruction of ECM. Overexpression of VSIG4 and MS4A4A can promote the M1-phenotype macrophages to transform into M2, and negatively regulate macrophage activation (39,40). High levels of CD33, ITGAM, CD14, CSF1R, NRP1, IL3RA, ITGAX and TGFB1 have been confirmed to be associated with the aggregation of MDSCs and DC cells in TME. MDSCs family and DC cells also accelerate the formation of an immunosuppressive TME via coordinating the response of immune inflammatory cell (41-43). Several studies revealed that CTSK has a higher stromal expression in tumour-associated fibroblasts and macrophages of invasive tumors compared to non-invasive carcinomas (44,45). In addition, according to a recent study, CTSK can accelerate metastasis in colorectal cancer via mediating TLR4-dependent M2 macrophage polarization with the help of gut microbiota (46).
GO analyses reflected that the BPs of DEGs are mainly enriched in ECM organization, protein complex subunit organization, endodermal cell differentiation, cellular response to cytokine stimulus, cellular protein modification process and platelet degranulation in our study. Additionally, the MFs are found to be in various biological binding like platelet-derived growth factor binding, integrin binding, collagen binding et al. The KEGG pathway enrichment revealed that the DEGs mainly act on the protein bio-utilization, ECM-receptor interaction, FA and PI3K-AKT signaling pathway. The ECM is an important part of cell micro-environment, whose structural function is essential to keep the normal activity of cell (47). However, recent studies have identified that the composition and mechanical properties of ECM take a significant part in the BPs associated with tumor development, such as escaping from apoptosis and regulating of cell growth, promoting tumor angiogenesis, gaining invasion and metastatic ability (48-51). Type IV collagen, one of the six subunits of which is encoded by COL4A2, is the most important component of the network structure of ECM and provides a tensile strength for underlying tissues, bounding diverse macromolecules and binding multiple cellular receptors. Similarly, evidences that COL4A2 might promote cell-adhesion, activate migration, and stimulate proliferation of different cell types has been reported, which are related to tumorigenesis (52-54). Actually, mutations in COL4A2 lead to its ectopic expression and eventually result in the occurrence of disease in the body. Even so, current researches on COL4A2 mutations mainly focus on the field of cerebrovascular diseases, but the details are still not quite clear as well as involving in the development of tumors (55). These, together with our results, suggest that CTSK and COL4A2 are indeed involved in matrix remodeling and enhance the invasion of tumor cells to a certain extent.
Based on the available evidences, we can attempt to explain the underlying molecular mechanisms of CTSK and COL4A2 involved in tumorigenesis. To become cancerous, cell has to break down the original connection between cell and cell, remodel cell-matrix adhesion site, develop along a pathway with the participation of enzymes secreted by ECM and finally undergo epithelial-mesenchymal transformation (EMT) (56,57). Aberrant EMT activation reduces the epithelial features of gastric epithelial cells and makes them obtain more characteristics of mesenchymal cells that tend to be dedifferentiated and more cancerous, and obtain capability to invade (58,59). Pathways like PI3K/AKT and TGF-β signaling pathway are also activated during the process of EMT, which further contributes to induce angiogenesis and inflammatory cell recruitment in TME (60,61). More simply, we can divide these complex processes into two steps: the first is remodeling of ECM that caused by the abnormal expression of hub genes such as COL4A2, CTSK et al. and the second is activation of EMT-cascade reactions that promote gastric epithelial cells towards mesenchymal cells, and acquire the capability of carcinogenesis. Meanwhile, in our findings, over-expression of CTSK can induce more immunocytes with inhibitive function such as M2 macrophages into TME and provide a suitable environment for cancerous cells growth, and allow cancer cells to escape the immune response in the end. Unfortunately, the formation of immunosuppressive microenvironment can further precipitate EMT process and tumor progression partly, and eventually lead to a poor prognosis of tumor patients. In summary, the most important role of CTSK and COL4A2 in cancers seems to be their contributions to the degradation of ECM-related proteins and the regulation of cytokines in TME.
However, many questions remain unanswered. Is CTSK involved in the regulation of tumor immunity as we have described? Is CTSK involved in regulating the tumor immune microenvironment continuous or intermittent? And what point in time does CTSK play a role in immunoregulation of normal gastric cells become cancerous? And what is the true relationship between CTSK, COL4A2 and EMT? Thus, further researches on these problems are needed by means of more experiments.
Conclusions
In conclusion, we identified two key genes (COL4A2 and CTSK) that could play a vital role in the pathogenesis of GC in Asian and act as the promising diagnostic and prognostic biomarkers in patients with GC probably via integrated bioinformatics analysis. CTSK could induce the formation of immunosuppressive TME and promote the immune escape of GC cells.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interests: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo.2020.03.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020;76:182-8. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet 2016;388:2654-64. [Crossref] [PubMed]
- Ott K, Vogelsang H, Mueller J, et al. Chromosomal instability rather than p53 mutation is associated with response to neoadjuvant cisplatin-based chemotherapy in gastric carcinoma. Clin Cancer Res 2003;9:2307-15. [PubMed]
- Sawaki A, Ohashi Y, Omuro Y, et al. Efficacy of trastuzumab in Japanese patients with HER2-positive advanced gastric or gastroesophageal junction cancer: a subgroup analysis of the Trastuzumab for Gastric Cancer (ToGA) study. Gastric Cancer 2012;15:313-22. [Crossref] [PubMed]
- Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449-56. [Crossref] [PubMed]
- Wang J, Sun Y, Bertagnolli MM. Comparison of gastric cancer survival between Caucasian and Asian patients treated in the United States: results from the Surveillance Epidemiology and End Results (SEER) database. Ann Surg Oncol 2015;22:2965-71. [Crossref] [PubMed]
- Fujita K, Yamamoto W, Endo S, et al. CYP2A6 and the plasma level of 5-chloro-2, 4-dihydroxypyridine are determinants of the pharmacokinetic variability of tegafur and 5-fluorouracil, respectively, in Japanese patients with cancer given S-1. Cancer Sci 2008;99:1049-54. [Crossref] [PubMed]
- Kim J, Sun CL, Mailey B, et al. Race and ethnicity correlate with survival in patients with gastric adenocarcinoma. Ann Oncol 2010;21:152-60. [Crossref] [PubMed]
- Jia F, Teer JK, Knepper TC, et al. Discordance of Somatic Mutations Between Asian and Caucasian Patient Populations with Gastric Cancer. Mol Diagn Ther 2017;21:179-85. [Crossref] [PubMed]
- Biagioni A, Skalamera I, Peri S, et al. Update on gastric cancer treatments and gene therapies. Cancer Metastasis Rev 2019;38:537-48. [Crossref] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 2017;33:2938-40. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-D613. [Crossref] [PubMed]
- Otasek D, Morris JH, Boucas J, et al. Cytoscape Automation: empowering workflow-based network analysis. Genome Biol 2019;20:185. [Crossref] [PubMed]
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. [Crossref] [PubMed]
- Nagy Á, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Siemers NO, Holloway JL, Chang H, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One 2017;12:e0179726. [Crossref] [PubMed]
- Danaher P, Warren S, Dennis L, et al. Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer 2017;5:18. [Crossref] [PubMed]
- Pan JH, Zhou H, Cooper L, et al. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front Immunol 2019;10:6. [Crossref] [PubMed]
- Li L, Zhu Z, Zhao Y, et al. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci Rep 2019;9:7827. [Crossref] [PubMed]
- Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol 2012;29:77-83. [Crossref] [PubMed]
- Li H, Yu B, Li J, et al. Characterization of differentially expressed genes involved in pathways associated with gastric cancer. PLoS One 2015;10:e0125013. [Crossref] [PubMed]
- Jin Y, He J, Du J, et al. Overexpression of HS6ST2 is associated with poor prognosis in patients with gastric cancer. Oncol Lett 2017;14:6191-7. [PubMed]
- He J, Jin Y, Chen Y, et al. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther 2016;9:6099-109. [Crossref] [PubMed]
- Qian Z, Zhu G, Tang L, et al. Whole genome gene copy number profiling of gastric cancer identifies PAK1 and KRAS gene amplification as therapy targets. Genes Chromosomes Cancer 2014;53:883-94. [Crossref] [PubMed]
- Oh SC, Sohn BH, Cheong JH, et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun 2018;9:1777. [Crossref] [PubMed]
- Lee J, Sohn I, Do IG, et al. Nanostring-based multigene assay to predict recurrence for gastric cancer patients after surgery. PLoS One 2014;9:e90133. [Crossref] [PubMed]
- Ashktorab H, Kupfer SS, Brim H, et al. Racial Disparity in Gastrointestinal Cancer Risk. Gastroenterology 2017;153:910-23. [Crossref] [PubMed]
- Le Gall C, Bellahcene A, Bonnelye E, et al. A cathepsin K inhibitor reduces breast cancer induced osteolysis and skeletal tumor burden. Cancer Res 2007;67:9894-902. [Crossref] [PubMed]
- Munari E, Cima L, Massari F, et al. Cathepsin K expression in castration-resistant prostate carcinoma: a therapeutical target for patients at risk for bone metastases. Int J Biol Markers 2017;32:e243-e247. [Crossref] [PubMed]
- Verbovšek U, Van Noorden CJ, Lah TT. Complexity of cancer protease biology: Cathepsin K expression and function in cancer progression. Semin Cancer Biol 2015;35:71-84. [Crossref] [PubMed]
- Duong LT, Wesolowski GA, Leung P, et al. Efficacy of a cathepsin K inhibitor in a preclinical model for prevention and treatment of breast cancer bone metastasis. Mol Cancer Ther 2014;13:2898-909. [Crossref] [PubMed]
- Ren G, Tian Q, An Y, et al. Coronin 3 promotes gastric cancer metastasis via the up-regulation of MMP-9 and cathepsin K. Mol Cancer 2012;11:67. [Crossref] [PubMed]
- Leusink FK, Koudounarakis E, Frank MH, et al. Cathepsin K associates with lymph node metastasis and poor prognosis in oral squamous cell carcinoma. BMC Cancer 2018;18:385. [Crossref] [PubMed]
- Wolf Y, Anderson AC, Kuchroo VK. TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol 2020;20:173-85. [Crossref] [PubMed]
- Li J, Diao B, Guo S, et al. VSIG4 inhibits proinflammatory macrophage activation by reprogramming mitochondrial pyruvate metabolism. Nat Commun 2017;8:1322. [Crossref] [PubMed]
- Sanyal R, Polyak MJ, Zuccolo J, et al. MS4A4A: a novel cell surface marker for M2 macrophages and plasma cells. Immunol Cell Biol 2017;95:611-9. [Crossref] [PubMed]
- Kitano Y, Okabe H, Yamashita YI, et al. Tumour-infiltrating inflammatory and immune cells in patients with extrahepatic cholangiocarcinoma. Br J Cancer 2018;118:171-80. [Crossref] [PubMed]
- Diakos CI, Charles KA, McMillan DC, et al. Cancer-related inflammation and treatment effectiveness. Lancet Oncol 2014;15:e493-503. [Crossref] [PubMed]
- Ino Y, Yamazaki-Itoh R, Shimada K, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer 2013;108:914-23. [Crossref] [PubMed]
- Kleer CG, Bloushtain-Qimron N, Chen YH, et al. Epithelial and stromal cathepsin K and CXCL14 expression in breast tumor progression. Clin Cancer Res 2008;14:5357-67. [Crossref] [PubMed]
- Rapa I, Volante M, Cappia S, et al. Cathepsin K is selectively expressed in the stroma of lung adenocarcinoma but not in bronchioloalveolar carcinoma. A useful marker of invasive growth. Am J Clin Pathol 2006;125:847-54. [Crossref] [PubMed]
- Li R, Zhou R, Wang H, et al. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ 2019;26:2447-63. [Crossref] [PubMed]
- Giussani M, Triulzi T, Sozzi G, et al. Tumor Extracellular Matrix Remodeling: New Perspectives as a Circulating Tool in the Diagnosis and Prognosis of Solid Tumors. Cells 2019;8:81. [Crossref] [PubMed]
- Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep 2014;15:1243-53. [Crossref] [PubMed]
- Eble JA, Niland S. The extracellular matrix in tumor progression and metastasis. Clin Exp Metastasis 2019;36:171-98. [Crossref] [PubMed]
- Poltavets V, Kochetkova M, Pitson SM, et al. The Role of the Extracellular Matrix and Its Molecular and Cellular Regulators in Cancer Cell Plasticity. Front Oncol 2018;8:431. [Crossref] [PubMed]
- Malik R, Lelkes PI, Cukierman E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol 2015;33:230-6. [Crossref] [PubMed]
- Boudko SP, Danylevych N, Hudson BG, et al. Basement membrane collagen IV: Isolation of functional domains. Methods Cell Biol 2018;143:171-85. [Crossref] [PubMed]
- Ohlund D, Lundin C, Ardnor B, et al. Type IV collagen is a tumour stroma-derived biomarker for pancreas cancer. Br J Cancer 2009;101:91-7. [Crossref] [PubMed]
- Brown CW, Brodsky AS, Freiman RN. Notch3 overexpression promotes anoikis resistance in epithelial ovarian cancer via upregulation of COL4A2. Mol Cancer Res 2015;13:78-85. [Crossref] [PubMed]
- Gunda B, Mine M, Kovacs T, et al. COL4A2 mutation causing adult onset recurrent intracerebral hemorrhage and leukoencephalopathy. J Neurol 2014;261:500-3. [Crossref] [PubMed]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006;7:131-42. [Crossref] [PubMed]
- Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 2009;28:15-33. [Crossref] [PubMed]
- Peng Z, Wang CX, Fang EH, et al. Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J Gastroenterol 2014;20:5403-10. [Crossref] [PubMed]
- Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol 2018;13:395-412. [Crossref] [PubMed]
- Thiery JP, Acloque H, Huang RYJ, et al. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009;139:871-90. [Crossref] [PubMed]
- Polivka J Jr, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther 2014;142:164-75. [Crossref] [PubMed]