Evaluation of phosphatidylinositol-3-kinase catalytic subunit (PIK3CA) and epidermal growth factor receptor (EGFR) gene mutations in pancreaticobiliary adenocarcinoma
Introduction
Epidermal growth factor receptor (EGFR)-mediated cell signaling, including the Ras/mitogen-activated protein kinase (MAPK) signaling pathway activation, plays an important role in angiogenesis, proliferation, and apoptosis (1,2). Tumors often exhibit alternations in receptor tyrosine kinases (TK) upstream of phosphatidylinositol-3-kinase (PI3K), the p110 catalytic subunit of PI3K (PIK3CA), the downstream kinase, Akt, and the negative regulator, PTEN (3). EGFR missense and deletion mutations were found in 13.4% of non-small cell lung cancer (NSCLC) patients, within exons 18 through 21 of the kinase domain (4). Lynch et al. reported in-frame deletions and amino acid substitutions, clustered in the region of the ATP-binding pocket of the TK domain, in eight of nine patients with gefitinib-responsive NSCLC (5). While EGFR mutations are characteristic for NSCLC, PIK3CA mutations are also identified in glioblastomas, colorectal cancer, gastric cancer, and breast cancer (3,6). EGFR is expressed by many epithelial tumor cells, including biliary and pancreatic cancers (7-9).
Inhibition of activated protein kinases through the use of targeted small molecule drugs (i.e., gefitinib and erlotinib) or antibody-based (i.e., cetuximab and panitumumab) strategies have emerged as an effective approach to cancer therapy (10-12). EGFR expression itself is not a definite predictor of response to EGFR TK inhibitors (13), however, EGFR mutations in NSCLC were found to predict sensitivity to gefitinib (4). Phase II studies have shown that TK inhibitors (TKI) induced response in over 70% of NSCLC patients harboring EGFR mutations (14).
Both pancreatic and biliary tract carcinoma are diagnosed at advanced stages when incurable, and outcomes even with surgery and chemotherapy, are poor (15-19). Combination of erlotinib and gemcitabine in advanced pancreatic cancer showed a modest increase in survival compared to gemcitabine alone, and resulted in the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMEA) approval for this regimen as first-line treatment of pancreatic cancer (20).
The objectives of this study were to determine the prevalence of EGFR and PI3K mutations in patients with pancreaticobiliary cancers. No studies had been reported at the time our research began of either EGFR or PIK3CA mutations in either disease. Several small reports have been published since, and this article will summarize the current literature in this field.
Materials and methods
Study population
This study was performed with approval of the Roswell Park Cancer Institute (RPCI) Institutional Review Board. The institutional pathology department reviewed all cases of pancreatic and biliary tract cancers following pancreatectomy diagnosed at RPCI over a period of five years between December 1, 1999, and November 30, 2004. All tumor blocks with adequate DNA for performing mutation analysis were selected for inclusion. Clinical data, including age, sex, ethnicity, and clinical stage, was obtained via chart review unblinded to mutation results. The samples were numbered consecutively to ensure patient confidentiality.
Histopathological evaluation
Twenty micron curls from tumor samples were examined with hematoxylin and eosin stain of the same area to ensure that the DNA is being extracted from a slice with maximal tumor and not normal tissue. The study examined mutational “hotspots” within the PIK3CA and EGFR genes based on reports by Pao et al. and Broderick et al. (4,6,21,22). The most frequently reported alterations in the PIK3CA gene in adult neoplasms are missense mutations in exon 9, which encodes a portion of the helical domain of the PIK3CA protein, and exon 20, which encodes the C-terminus of p110α catalytic subunit. PIK3CA gene mutations are believed to be activating mutations, and NIH3T3 cells transfected with H1047R (exon 20) mutant p110α constructs have increased lipid kinase activity as compared to cells transfected with wild-type p110α (21).
Mutational analysis was also performed for exons 18-21 of the EGFR gene that encode the protein TK domain of the EGFR protein. In-frame deletions in a GXGXXG motif (exon 19) as well as the missense mutations G719S (exon 18) and L858R (exon 21) of the EGFR gene associated with response to gefitinib were studied (Figure 1).
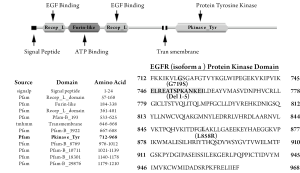
DNA extraction
A portion of snap frozen tumor biopsies from each patient were homogenized individually in TE buffer. Genomic DNA was extracted from each sample using a standard phenol/chloroform protocol, and the DNA quality was assessed by both spectrophotometry and visualization on an ethidium bromide stained agarose gel. Working dilutions of 50 ng/µL were prepared for each sample and DNA samples were stored at 4 °C.
PCR and direct sequencing
Primers sequences for exons 9 and 20 of the PIK3CA gene were designed using NCBI (
Statistical analysis
A description of the presence/absence of these known mutations in each of the two malignancies was tabulated and reported as a percentage of the total number of samples screened.
Review of literature
For the literature review, the electronic databases PubMed and MEDLINE, as well as the Cochrane library and the American Society of Clinical Oncology (ASCO) abstracts were searched using the key words pancreatic, pancreas, biliary, cholangiocarcinoma, cancer, EGFR, PIK3CA, and mutation, in all possible combinations, limited to humans and English-language studies. All articles published by the year 2011 were included. The authors adhered to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.
Results
Demographics and clinicopathological features of study population
Table 1 summarizes the clinicopathological findings for the 30 patients included in this study. The number of males was equal to the number of females. The median age was 63.8 (range, 46 to 84) years. Most of the patients were Caucasians, and over 86% (26 patients) were diagnosed with pancreatic adenocarcinoma.
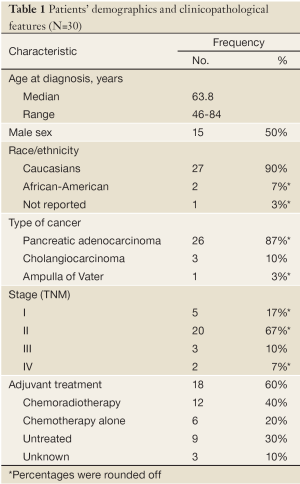
Full table
Twenty patients (67%) had a TNM stage II disease, 5 patients (17%) had a stage I disease, and 5 patients (17%) had advanced disease (stage III or IV). The patients who were found to have advanced disease had a clinically resectable tumor, yet final pathological report showed a T4 tumor and/or micrometastatic disease.
One patient was treated with neoadjuvant chemoradio-therapy with doxorubicin, 5-fluorouracil (5-FU), and cisplatin. Eighteen (60%) patients were treated with adjuvant therapy, two-third of them with a combination of chemoradiotherapy and the remaining third with chemotherapy alone. Ten patients were treated with 5-FU or capecitabine, four patients received gemcitabine, and two patients received a combination of both. One patient was treated with 5-FU in combination with streptozotocin, mitomycin, and leucovorin.
Subsequent therapies at the time of metastases detection were gemcitabine, oxaliplatin, or taxane based. Only one patient received gemcitabine in combination with erlotinib (Tarceva®, OSI Pharmaceuticals, LLC).
Sequence information
No mutations were identified in either exons 9 and 20 of the PIK3CA gene or in exons 18-21 of the EGFR gene in the 30 pancreaticobiliary tumors that were analyzed. One single nucleotide polymorphism (SNP), rs45455192, located in intron 8 of the PIK3CA gene was identified in a single tumor sample. This particular SNP has no reported frequency data. No unusual clinical findings were seen in the patient who provided this sample (Figure 2A).
An unreported variant in intron 18 of the EGFR gene was identified in one tumor sample (Figure 2B). This variant (C>T) occurred sixteen base pairs immediately following exon 18 (IVS18+16) and was near a reported SNP (rs17337107). This variant was queried as a possible splice site mutation, but no evidence for this change adversely affecting splicing was identified using splice site prediction software.

Review of literature
Review of the literature yielded thirteen relevant articles and abstracts relating to EGFR and/or PIK3CA mutations in human pancreatic adenocarcinoma. This is summarized in Table 2, including the results of this current study. Overall only 7 EGFR mutations were found among 347 pancreatic cancers (2%). Kwak et al. demonstrated disease stabilization with EGFR inhibition (erlotinib with capecitabine) in 5 out of 55 cases, including both (2/2) pancreatic cancers with EGFR mutations (24). PIK3CA mutations were identified in 2 out of 56 cases (3.6%) of pancreatic adenocarcinoma. Jimeno et al. found that 2 of 10 human tumors were sensitive to EGFR inhibition, including the single (1/1) pancreatic cancer with PIK3CA mutation (30).
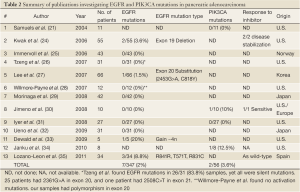
Full table
Similar search yielded eight articles and abstracts that investigated biliary tract carcinomas. The summary of these publications is presented in Table 3, including the results of this current study. A total of 19 EGFR mutations (10.5%) and 18 PIK3CA mutations (11.7%) were found in 180 and 154 biliary tumors, respectively. The latter percentage was influenced by the presence of PIK3CA mutations in one third of Chinese study population (8).
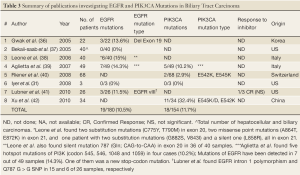
Full table
Discussion
EGFR activation influences different intertwining signaling pathways, including Ras/MAPK, phospholipase C, PI3K/Akt, signal transducer and activator of transcription, and Src/FAK pathways (43). EGFR is expressed by pancreatic tumor cells (7), and has been associated with lymph node involvement, metastasis and disease recurrence (44,45), and overall worse prognosis (46). High EGFR expression has been reported also in biliary cancer (8,9,47,48). Tan et al. demonstrated that activation of EGFR is closely involved in cell dissociation in pancreatic cancer through activating MEK/ERK signaling pathway (49). Cytoplasmic overexpression of EGFR plays a significant role in the progression of pancreatic ductal adenocarcinoma, especially in the invasion and acquisition of aggressive clinical behavior (46). EGFR also contributes greatly to cholangiocarcinoma progression, associated with lymph node metastasis, aberrant p53 expression, proliferating activity, and carcinoma differentiation (50). EGFR is activated by bile acids and functions to induce COX-2 expression by an MAPK cascade that may contribute to progression of cholangiocarcinomas (51). Paez et al. searched for somatic genetic alterations in NSCLC specimens from Japan and the US by examining exons 2 through 25 of EGFR. They found missense and deletion mutations of EGFR in 13.4% of tumors, all within exons 18 through 21 of the kinase domain. The EGFR mutations were more frequent in adenocarcinomas, females, and Japanese patients (25% mutation prevalence vs. 1.6% in Americans) (4). The common EGFR mutations in NSCLC are exon 19 deletions and the L858R point mutation in exon 21 (52).
PI3K activation has been shown to play an important role in cell survival signaling in a number of cell types (53). The classic mode of PI3K activation involves its binding to phosphorylated tyrosine residues of receptor tyrosine kinases, such as EGF and the platelet-derived growth factor receptor. PI3K/Akt signaling promotes small-cell lung carcinoma (SCLC) growth, survival, and chemotherapy resistance (54). The PI3K pathway is activated in multiple advanced cancers through inactivation of the PTEN tumor suppressor gene (6). Systematic analysis of kinase genes has identified mutations in PI3K p110 catalytic subunit gene PIK3CA in human cancers (3,21,23). These missense mutations, H1047R, E545K and E542K, cluster in two conserved gene locations, and are mutations that confer constitutive kinase activity (21,55). PIK3CA gene is also amplified at high frequencies in squamous cell lung carcinoma, head and neck, gastric, and cervical cancers (56).
Carcinoma of the pancreas is the fourth leading cause of cancer mortality in the U.S. Unfortunately its survival has not improved substantially over the past thirty years, with median survival in the metastatic stage of six months (16,17).
TK inhibitors have been shown to improve the outcome in patients with lung and pancreatic cancers (43). EGFR overexpression by immunohistochemistry is significantly higher in pancreatic tumor cells when compared to normal pancreatic cells (7). Erlotinib is a human EGFR type 1 (HER1)/EGFR TK inhibitor. As a single first or second line agent pancreatic disease control for more than eight weeks was achieved in 20% of patients (57). The drug was approved by the FDA initially for advanced NSCLC, and in 2005 for advanced pancreatic cancer combined with gemcitabine (58). So far only erlotinib has been shown to improve survival in pancreatic adenocarcinoma, with one-year survival of 23% in the erlotinib group compared to 17% with gemcitabine monotherapy (20).
Cholangiocarcinoma is a rare and aggressive tumor that is similar to pancreatic adenocarcinoma, both in histological features and in clinical outcome (18,59,60). Philip et al. reported EGFR expression rate of 81% in patients with unresectable or metastatic biliary disease. Following anti-EGFR therapy, 17 percent of patients were progression free at six months; however, EGFR expression in baseline tumor specimens did not correlate with treatment benefit (48,61).
Gefitinib (Iressa), another EGFR inhibitor, inhibits pancreatic cancer cell growth through EGFR-dependent pathways and delays anchorage-independent growth and invasiveness (62). It was approved in Japan and the US for the treatment of NSCLC. The original rationale for its use was the observation that EGFR is abundantly expressed in lung carcinoma tissue in comparison to adjacent normal lung (63). However, EGFR expression as detected by immunohistochemistry is not an effective predictor of response to gefitinib (13). Presence of specific mutations in the EGFR gene has been shown to correlate with clinical response to the EGFR inhibitor gefitinib in patients with advanced non small cell lung cancer (4).
Erlotinib has been shown to have efficacy in pancreatic and biliary cancers, yet there was no published data on predictive value or prevalence of the abovementioned mutations in these tumor types, therefore this study was undertaken. The study failed to identify mutations in either PIK3CA or EGFR genes for any of the thirty pancreaticobiliary tumor samples that were analyzed. It did identify one synonymous SNP (rs1050171) in the coding region of EGFR, and a previously unreported change, suspected to be a SNP, in intron 18 of EGFR (IVS18+15, C>T). The main limitation of our study is the small population size for pancreatic cancer and biliary tract cancer. Therefore, we conducted a review of the literature to explore the total number of patients and mutation detection. The review showed an EGFR mutation rate of 2% and 10.5% in pancreatic and biliary tract carcinomas, respectively. PIK3CA mutations were demonstrated in 3.6% and 11.7% of pancreatic and biliary tract carcinomas, respectively. This pooled data from the literature is in concordance with our study, showing similar rates in pancreatic adenocarcinoma. The prevalence of EGFR and PIK3CA mutations reported in the literature for pancreatic cancer was less than 5%. This finding may explain in part why erlotinib provides only a modest improvement in survival, as many other factors might play a role in the prognosis. Another limitation results from the inclusion of a single patient who received neoadjuvant therapy, thus the desmoplastic component of the tumor might have interfered with sequencing.
Genome-wide analysis is being utilized to identify mutations that might have an importance in diagnosis, prognosis, and treatment of pancreatic cancer. Harada et al. found frequent dysregulation of SKAP2/SCAP2 gene (7p15.2) in pancreatic cancer (64). Vincent et al. found numerous target genes that were hypermethylated and silenced or hypomethylated and overexpressed (65), while Jones et al. reported that pancreatic cancers have approximately 63 genetic alterations, mainly point mutations, which affect cellular signaling pathways (66).
In contrast to pancreatic cancer, biliary tract cancer had a higher prevalence of both EGFR and PIK3CA mutations, slightly over 10%, a value similar to that of EGFR mutation in NSCLC (4). Xu et al. reported that one third of Chinese patients with cholangiocarcinoma had PIK3CA mutations (42). This relatively high prevalence rate in Asian population might explain the varied response to treatment in different populations. Despite the fact that biliary tract cancer and pancreatic cancer share similar clinicopathological characteristics, the variation in EGFR and PIK3CA mutation rates might indicate that they have different pathophysiology. This research provides the background for designing future correlative prospective trials with EGFR inhibitors. It highlights the importance of studying the biology of each tumor due to their noted variability. It is conceivable that these mutational studies will improve our understanding of tumor biology, and may refine targeted approaches in these dismal diseases. Surgical attention must be given to the creation of fresh frozen specimen banks, as sensitivity of mutation detection may be higher in fresh frozen rather than paraffin embedded specimens. The role of other mutations, such as K-RAS, predictive of response to EGFR inhibition with monoclonal antibodies in colon cancer, needs further investigation in these diseases. Future targeted therapy should take into account treatment regimens- as monotherapy or in combination with current chemotherapy.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Wieduwilt MJ, Moasser MM. The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell Mol Life Sci 2008;65:1566-84.
- Chapman MS, Miner JN. Novel mitogen-activated protein kinase kinase inhibitors. Expert Opin Investig Drugs 2011;20:209-20.
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene 2008;27:5497-510.
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500.
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39.
- Broderick DK, Di C, Parrett TJ, et al. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res 2004;64:5048-50.
- Muslimov GF. Role of epidermal growth factor gene in the development of pancreatic cancer and efficiency of inhibitors of this gene in the treatment of pancreatic carcinoma. Bull Exp Biol Med 2008;145:535-8.
- Nakazawa K, Dobashi Y, Suzuki S, et al. Amplification and overexpression of c-erbB-2, epidermal growth factor receptor, and c-met in biliary tract cancers. J Pathol 2005;206:356-65.
- Harder J, Waiz O, Otto F, et al. EGFR and HER2 expression in advanced biliary tract cancer. World J Gastroenterol 2009;15:4511-7.
- Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 2002;347:472-80.
- Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med 2001;344:1038-42.
- Krempien R, Muenter MW, Huber PE, et al. Randomized phase II--study evaluating EGFR targeting therapy with cetuximab in combination with radiotherapy and chemotherapy for patients with locally advanced pancreatic cancer--PARC: study protocol [ISRCTN56652283]. BMC Cancer 2005;5:131.
- Renee B, Mark K, Michael W, et al. O-242 Gefitinib (‘Iressa’, ZD1839) monotherapy for pretreated advanced non-small-cell lung cancer in IDEAL 1 and 2: tumor response is not clinically relevantly predictable from tumor EGFR membrane staining alone. Lung cancer 2003;41:S71.
- Rosell R, Moran T, Cardenal F, et al. Predictive biomarkers in the management of EGFR mutant lung cancer. Ann N Y Acad Sci 2010;1210:45-52.
- Burtness B. Her signaling in pancreatic cancer. Expert Opin Biol Ther 2007;7:823-9.
- Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300.
- Altekruse SF, Kosary CL, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2007. Bethesda, Md. National Cancer Institute 2010.
- Patel T. Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology 2001;33:1353-7.
- Boku N. Emerging treatment with systemic chemotherapy and targeted agents for biliary cancers. Curr Opin Investig Drugs 2010;11:653-60.
- Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol 2007;25:1960-6.
- Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004;304:554.
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11.
- Bardelli A, Parsons DW, Silliman N, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science 2003;300:949.
- Kwak EL, Jankowski J, Thayer SP, et al. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin Cancer Res 2006;12:4283-7.
- Immervoll H, Hoem D, Kugarajh K, et al. Molecular analysis of the EGFR-RAS-RAF pathway in pancreatic ductal adenocarcinomas: lack of mutations in the BRAF and EGFR genes. Virchows Arch 2006;448:788-96.
- Tzeng CW, Frolov A, Frolova N, et al. Epidermal growth factor receptor (EGFR) is highly conserved in pancreatic cancer. Surgery 2007;141:464-9.
- Lee J, Jang KT, Ki CS, et al. Impact of epidermal growth factor receptor (EGFR) kinase mutations, EGFR gene amplifications, and KRAS mutations on survival of pancreatic adenocarcinoma. Cancer 2007;109:1561-9.
- Willmore-Payne C, Volmar KE, Huening MA, et al. Molecular diagnostic testing as an adjunct to morphologic evaluation of pancreatic ductal system brushings: potential augmentation for diagnostic sensitivity. Diagn Cytopathol 2007;35:218-24.
- Morinaga S SN, Wada H, Shiozawa M, et al. Epidermal growth factor receptor (EGFR) status: Mutation and protein expression, and clinico-pathology of pancreatic ductal adenocarcinoma. 2008 ASCO Gastrointestinal Cancers Symposium. Abstract No. 185.
- Jimeno A, Tan AC, Coffa J, et al. Coordinated epidermal growth factor receptor pathway gene overexpression predicts epidermal growth factor receptor inhibitor sensitivity in pancreatic cancer. Cancer Res 2008;68:2841-9.
- Iyer RV RM, Bharthuar A, Levea C, et al. Evaluation of phosphatidyl inositol 3-kinase catalytic subunit (PI3KCA) and epidermal growth factor receptor (EGFR) gene mutations in pancreatic adenocarcinoma. 2009 Gastrointestinal Cancers Symposium Proceedings. Abstract No. 173.
- Ueno M, Ohkawa S, Sakamoto Y, et al. The analysis of EGFR expression and EGFR mutations in advanced pancreatic cancer. J Clin Oncol 2009;27: e15629.
- Dewald GW, Smyrk TC, Thorland EC, et al. Fluorescence in situ hybridization to visualize genetic abnormalities in interphase cells of acinar cell carcinoma, ductal adenocarcinoma, and islet cell carcinoma of the pancreas. Mayo Clin Proc 2009;84:801-10.
- Janku F, Tsimberidou AM, Garrido-Laguna I, et al. PIK3CA, KRAS, and BRAF mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. J Clin Oncol (Meeting Abstracts) 2010;28:2583.
- Lozano-Leon A, Perez-Quintela BV, Iglesias-García J, et al. Clinical relevance of epidermal growth factor receptor (EGFR) alterations in human pancreatic tumors. Oncol Rep 2011;26:315-20.
- Gwak GY, Yoon JH, Shin CM, et al. Detection of response-predicting mutations in the kinase domain of the epidermal growth factor receptor gene in cholangiocarcinomas. J Cancer Res Clin Oncol 2005;131:649-52.
- Bekaii-Saab TS ST, Williams N, Frankel W, et al. Gain-of-function somatic mutations affecting the catalytic domain of EGFR as described in non-small cell lung carcinomas are not present in hepatobiliary tumors. 2005 Gastrointestinal Cancers Symposium Proceedings. Abstract No. 123.
- Leone F, Cavalloni G, Pignochino Y, et al. Somatic mutations of epidermal growth factor receptor in bile duct and gallbladder carcinoma. Clin Cancer Res 2006;12:1680-5.
- Aglietta M, Pignochino Y, Cavalloni G, et al. Somatic mutations of EGFR signal transducers and expression of tumor suppressor PTEN in biliary tract carcinoma. J Clin Oncol (Meeting Abstracts) 2007;25:4582.
- Riener MO, Bawohl M, Clavien PA, et al. Rare PIK3CA hotspot mutations in carcinomas of the biliary tract. Genes Chromosomes Cancer 2008;47:363-7.
- Lubner SJ, Mahoney MR, Kolesar JL, et al. Report of a multicenter phase II trial testing a combination of biweekly bevacizumab and daily erlotinib in patients with unresectable biliary cancer: a phase II Consortium study. J Clin Oncol 2010;28:3491-7.
- Xu RF, Sun JP, Zhang SR, et al. KRAS and PIK3CA but not BRAF genes are frequently mutated in Chinese cholangiocarcinoma patients. Biomed Pharmacother 2011;65:22-6.
- Laurent-Puig P, Lievre A, Blons H. Mutations and response to epidermal growth factor receptor inhibitors. Clin Cancer Res 2009;15:1133-9.
- Tobita K, Kijima H, Dowaki S, et al. Epidermal growth factor receptor expression in human pancreatic cancer: Significance for liver metastasis. Int J Mol Med 2003;11:305-9.
- Pryczynicz A, Guzińska-Ustymowicz K, Czyzewska J, et al. Expression of epidermal growth factors and apoptosis markers in pancreatic ductal adenocarcinoma. Folia Histochem Cytobiol 2009;47:667-71.
- Ueda S, Ogata S, Tsuda H, et al. The correlation between cytoplasmic overexpression of epidermal growth factor receptor and tumor aggressiveness: poor prognosis in patients with pancreatic ductal adenocarcinoma. Pancreas 2004;29:e1-8.
- Lee CS, Pirdas A. Epidermal growth factor receptor immunoreactivity in gallbladder and extrahepatic biliary tract tumours. Pathol Res Pract 1995;191:1087-91.
- Philip PA, Mahoney MR, Allmer C, et al. Phase II study of erlotinib in patients with advanced biliary cancer. J Clin Oncol 2006;24:3069-74.
- Tan X, Egami H, Ishikawa S, et al. Relationship between activation of epidermal growth factor receptor and cell dissociation in pancreatic cancer. Int J Oncol 2004;25:1303-9.
- Ito Y, Takeda T, Sasaki Y, et al. Expression and clinical significance of the erbB family in intrahepatic cholangiocellular carcinoma. Pathol Res Pract 2001;197:95-100.
- Yoon JH, Higuchi H, Werneburg NW, et al. Bile acids induce cyclooxygenase-2 expression via the epidermal growth factor receptor in a human cholangiocarcinoma cell line. Gastroenterology 2002;122:985-93.
- Jänne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol 2005;23:3227-34.
- Reusch HP, Zimmermann S, Schaefer M, et al. Regulation of Raf by Akt controls growth and differentiation in vascular smooth muscle cells. J Biol Chem 2001;276:33630-7.
- Krystal GW, Sulanke G, Litz J. Inhibition of phosphatidylinositol 3-kinase-Akt signaling blocks growth, promotes apoptosis, and enhances sensitivity of small cell lung cancer cells to chemotherapy. Mol Cancer Ther 2002;1:913-22.
- Samuels Y, Diaz LA Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 2005;7:561-73.
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 2006;7:606-19.
- Iyer R, Khushalani N, Tan W, et al. A phase II study of erlotinib in patients (pts) with advanced pancreatic cancer (APC) who are refractory to gemcitabine (G). 2010 ASCO Gastrointestinal Cancers Symposium. Abstract No. 258.
- Senderowicz AM, Johnson JR, Sridhara R, et al. Erlotinib/gemcitabine for first-line treatment of locally advanced or metastatic adenocarcinoma of the pancreas. Oncology (Williston Park) 2007;21:1696-706; discussion 1706-9, 1712, 1715.
- Wiedmann MW, Mössner J. Molecular targeted therapy of biliary tract cancer--results of the first clinical studies. Curr Drug Targets 2010;11:834-50.
- Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis 2004;24:115-25.
- Philip PA, Benedetti J, Corless CL, et al. Phase III study comparing gemcitabine plus cetuximab versus gemcitabine in patients with advanced pancreatic adenocarcinoma: Southwest Oncology Group-directed intergroup trial S0205. J Clin Oncol 2010;28:3605-10.
- Li J, Kleeff J, Giese N, et al. Gefitinib (‘Iressa’, ZD1839), a selective epidermal growth factor receptor tyrosine kinase inhibitor, inhibits pancreatic cancer cell growth, invasion, and colony formation. Int J Oncol 2004;25:203-10.
- Rusch V, Baselga J, Cordon-Cardo C, et al. Differential expression of the epidermal growth factor receptor and its ligands in primary non-small cell lung cancers and adjacent benign lung. Cancer Res 1993;53:2379-85.
- Harada T, Chelala C, Bhakta V, et al. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene 2008;27:1951-60.
- Vincent A, Omura N, Hong SM, et al. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res 2011;17:4341-54.
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801-6.


