Identification of potential biomarkers and pathways in ulcerative colitis with combined public mRNA and miRNA expression microarray data analysis
Introduction
Ulcerative colitis (UC), a subtype of inflammatory bowel disease (IBD), is a chronic, relapsing and non-specific inflammatory disease, which is confined to the mucosa and submucosa of the rectum or colon. It is characterized by two periods of active disease and remission. The typical clinical manifestations are diarrhea, purulent stool and abdominal pain (1).
UC afflicts millions of people worldwide. The highest incidences of UC in North America and Northern Europe were 6–15.6 and 10–20.3 cases, respectively, per 100,000 annually (2). A recent review analysis of 44 studies that included 31,287 Asian patients found a 0.85% prevalence rates of UC (3). Additionally, patient suffering from UC are at high risk of developing colorectal cancer (CRC) (4). UC has brought significant personal and societal burden. Furthermore, active UC can decrease the physical and mental quality of life and increase psychological distress.
MicroRNAs (miRNAs), a group of small non-coding RNA molecules, act as key post-transcriptional regulators of gene expression. The dysregulated miRNA expression can drive the onset of various diseases and even contribute to oncogenesis or tumor progression. miRNAs have cytoprotective effect on tissue as protectomiRs (5), and play a pivotal role in the polarization of tumor-related macrophages in inflammatory microenvironment (6). Moreover, miRNAs function of inflammation and autoimmunity in regulating the pathogenesis of UC (7). Mounting gene expression studies performed in UC both in colonic mucosa and peripheral blood have proposed various hypothesis, that is, cell adhesion, immune response and tissue remodeling (8). Numerous genes and pathways are correlated with the genesis and progression of UC. For example, high expression levels of Caspase 1 (CASP1) and lysozyme (LYZ) have been found in UC group (9). Various genes, such as CD177 Molecule (CD177), G protein-coupled receptor 84 (GPR84) (10), interleukin 8 (IL-8), matrix metallopeptidase 9 (MMP-9) (11), and 8-Oxoguanine DNA glycosylase (OGG1) (12), participate in molecular pathways of UC. Also, various signaling pathways, such as nuclear transcription factor kappa B (NF-κB) pathway (13), nuclear factor erythroid 2-related factor 2 (Nrf2) pathway (14), and JAK/STAT signaling pathway (15) are related to progress of UC. However, knowledge of the genome scale of miRNAs and their target genes, as well as the potential biological function of UC, remains limited.
In this study, we retrieved datasets of mRNA and miRNA expression microarrays from the Gene Expression Omnibus (GEO), and we identified a group of key miRNAs and potentially target genes involved in UC by using bioinformatics analysis. In addition, the most significantly expressed miRNAs and their target genes were selected and conducted for preliminary validation by real-time qPCR (qRT-PCR). The study aimed to suggest miRNA signatures useful for active UC detection and diagnosis, as well as explore the underlying pathogenesis by identifying potential miRNA-targeted mRNAs at the molecular level.
Methods
Collection and inclusion criteria of studies
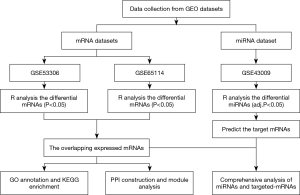
The mRNA microarray expression profile datasets were retrieved and downloaded from the GEO database (available online: http://www.ncbi.nlm.nih.gov/geo) by searching the following key words: ‘RNA’, ‘active ulcerative colitis’, and ‘Homo sapiens’ (organism). The inclusion criteria were as follows: (I) colon tissues from adult patients with active UC (not cells); (II) samples in UC group without receiving any interventions or treatments; and (III) both numbers of UC and health control sample ≥12. Besides, the miRNA profile datasets were searched by using the keywords: ‘miRNA’, ‘active ulcerative colitis’, and ‘Homo sapiens’ (organism) according to the following criteria: (I) colon tissues from active UC patients (not cells) and (II) samples in UC group without receiving any interventions or treatments. After screening, two mRNA expression datasets [GSE53306 (16) and GSE65114] were selected, and one miRNA expression dataset (GSE43009) was obtained for analysis. The workflow of data-processing and analysis is presented in Figure 1.
Microarray data
In this study, the platform for GSE53306 was based on the GPL14951 Illumina HumanHT-12 WG-DASL V4.0 R2 expression beadchip, which consisted of 16 active UC samples and 12 controls. The platform for GSE65114 was GPL16686 (HuGene-2_0-st) Affymetrix human gene 2.0 ST array, which included 16 active UC samples and 12 controls. The platform for GSE43009 was based on GPL16384 Affymetrix multispecies miRNA-3 array, which consisted of five controls and five UC samples.
Data-processing and differentially expressed genes (DEGs)/differentially expressed miRNAs (DEMI) identification
The raw data were downloaded from the GEO database and then normalized and standardized by using the R software package. Gene differential expression analysis was conducted through the limma packages in the Bioconductor package (17) (available online: http://www.bioconductor.org/). The heat maps of the two mRNA datasets were mapped by using the gplots package in R to visualize the expression values of genes in the different samples. When we selected the DEGs, P<0.05 and |log FC| >1 were considered as the cutoff values, where FC is fold change. Significant DEMIs were screened by meeting both adj. P<0.05 and |log FC| >1. The Venny online tool (available online: http://bioinfogp.cnb.csic.es/tools/venny) was used to identify DEGs across the two mRNAs datasets. The identified DEMIs were preserved for further bioinformatics analysis.
Functional enrichment analysis of DEGs
Gene ontology (GO) enrichment analysis, which is used for annotating genes, was conducted to determine significantly regulated functions, that is, biological processes (BP), cellular component (CC) and molecular function (MF). The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed for presenting the systematic analysis, annotation, and visualization of gene functions. Both GO enrichment and KEGG pathway analysis were conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (available online: http://david.abcc.ncifcrf.gov/) to identify the biological significance of genes, when P<0.05 was considered statistically significant.
Protein-protein interaction (PPI) network construction and module selection
The PPI network of DEGs was mapped using the Search Tool for the Retrieval of Interacting Genes (STRING, available online: http://string.embl.de/) to evaluate the interactions of protein pairs, with confidence score >0.4 defined as cutoff criterion. The integrated regulatory networks were then visualized by Cytoscape (18). Finally, the plug-in Molecular Complex Detection (MCODE) was applied to screen the modules of PPI network.
Prediction of miRNA targets
Relevant miRNA targets were predicted using miRWalk (available online: http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/), which is a comprehensive atlas of predicted and validated miRNA-target interactions. The potential targets of miRNA were identified by at least four programs.
Animals and experimental design
Twenty male SD rat (180±20 g) aging 8–10 weeks were purchased from the Qinglongshan Experimental Animal Breeding Farm (Nanjing, China). Animals were housed in a standard condition with 22±2 °C ambient temperature at 12 h light/darkness. The rats with unlimited access to standard rat chow were randomly assigned into two groups (ten rats per group). The acute colitis rats were induced by giving drink water containing 3.5% (w/v) dextran sulfate sodium (DSS) (Sigma-Aldrich, USA), while the controls were given DSS-free drinking water. After 10 days, all rats were sacrificed under isoflurane anesthesia, and the colon tissues were harvested for following analysis. All animal protocols carried out were in compliance with the Chinese Guidelines of Accommodation and Care for Animals formulated and under the approval of Nanjing University of Chinese Medicine.
RNA extraction and qRT-PCR
According to the manufacturer’s instruction, total RNA of colon tissues in both UC groups and controls were extracted using the TRIzol Reagent (Invitrogen, California, USA). The primers were obtained from Genscript, and the sequences were presented in Table S1. The mRNA and miRNA expression was normalized against GAPDH and U6 expression, respectively.

Full table
Hematoxylin and eosin (HE) staining
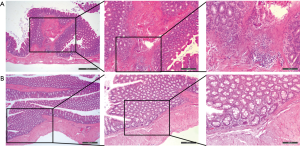
The colon tissues were sliced into 4 µm thick section after fixation, dehydration and embedding and then stained with hematoxylin and eosin. Three random sections of each tissue were imaged under the microscope (Leica, German).
Statistical analyses
Statistical analyses were conducted using SPSS 19.0 software. All data were expressed as the mean ± standard deviation (SD) and analyzed using Graph Pad Prism (Version 5.0, Inc., CA, USA). Statistical comparisons within two groups were made by unpaired Student t-tests. And P value less than 0.05 was considered significant.
Results
Identification of DEGs
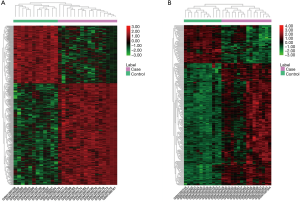
The two microarray datasets GSE53306 and GSE65114 were normalized, as shown in Figure 2. A total of 1,052 DEGs were screened from the GSE53306 dataset. Additionally, 296 DEGs were identified from the GSE65114 dataset. The hierarchical cluster heatmaps of DEGs in both datasets are presented in Figure 3. And 79 genes were differentially expressed in both two datasets.
Functional and pathway enrichment analyses of DEGs
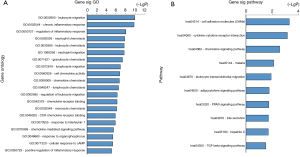
The 79 overlapping genes above were uploaded to the DAVID online analysis tool for functional assignment and pathway enrichment with a P value <0.05. We identified the top 10 most enriched GOs, including leukocyte migration, chronic inflammatory response, regulation of inflammatory response, neutrophil chemotaxis, leukocyte chemotaxis, neutrophil migration, granulocyte chemotaxis, lymphocyte migration, cell chemotaxis, and chemokine activity, as represented in Figure 4A and Table 1. Moreover, functional enrichment analysis based on GO terms was conducted significantly participated in UC-related pathways, such as cell adhesion molecules (CAMs), cytokine-cytokine receptor interaction, chemokine signaling pathway, adipocytokine signaling pathway, peroxisome proliferators-activated receptor (PPAR) and transforming growth factor beta (TGF-β) signaling pathway (Figure 4B and Table 2).

Full table

Full table
PPI network and module analysis
To further mine the UC-associated genes, we mapped PPI network analysis by using the STRING database, which was constructed by 79 nodes and 55 edges (Figure 5A) after removing the disconnected nodes with confidence score >0.4. We found several hub genes, such as metallopeptidase inhibitor 1 (TIMP1), C-X-C motif chemokine ligand 10 (CXCL10), C-X-C motif chemokine ligand 13 (CXCL13), C-C motif chemokine ligand 11 (CCL11), C-C motif chemokine ligand 19 (CCL19), and selectin L (SELL).
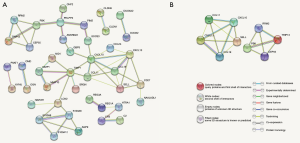
Subsequently, the 55 obtained PPI pairs underwent analysis by using Cytoscape to depict the complex relationship. Moreover, two clusters were identified from plug-in DCOME, which consisted of nine genes (Figure 5B), SELL, CCL19, CXCL13, CXCL10, CCL11, ribonucleotide reductase regulatory subunit M2 (RRM2), PDZ binding kinase (PBK), centrosomal protein 55 (CEP55), and thyroid hormone receptor interactor 13 (TRIP13). Finally, we found that these genes were enriched in the immune response by DAVID.
Prediction of DEMIs and identification of potential target genes
The profiling dataset GSE43009 was used to identify the DEMIs. After data preprocessing, we eliminated one group in the UC sample and control sample due to poor hierarchical cluster. A total of 47 DEMIs were identified. Next, by using miRWalk database, the predicted targets of miRNAs were obtained. The overlapping mRNAs predicted by miRWalk and the DEGs identified above are presented in Table 3. We found that miR-92b and miR-625 were the most significant miRNAs. PTGIS was predicted as the potential target of four miRNAs, namely, miR-1228, miR-1268, miR-1231 and miR-92b. DES was identified as potential target of three miRNAs, namely, miR-939, miR-1268 and miR-1226. UNC13D was found as potential target of miR-939, miR-1268 and miR-1908. Moreover, seven genes (including MMP10, DPP10, and PCK1) were potentially targeted by miR-92b, whereas seven mRNAs (including CCL11) were potentially targeted by miR-625.

Full table
Verification of miRNA and the target mRNAs by qRT-PCR
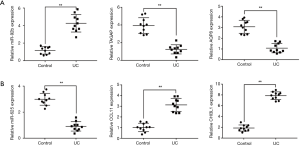
To demonstrate the miRNAs and the target mRNAs we identified, we chosen the most significantly up/down-expressed miRNA (miR-92b and miR-625), and we selected two of their corresponding target mRNAs (AQP8 and TAGAP, CCL11 and CHI3L1) based on the results of bioinformatics analysis and previous literatures to validate in colon tissues on rodent UC models. The HE staining of DSS-induced UC and control (Figure 6) suggested the successful model preparation. The results of qRT-PCR indicated that the miRNAs and the corresponding mRNAs were significantly expressed (P<0.05, Table 4 and Figure 7) in UC tissues compared with the controls, which were consistent with the results of microarray analysis.

Full table
Conclusions
In the present study, a total of 79 DEGs and 47 DEMIs were identified from the GEO datasets, which might provide initial evidence that these genes and miRNAs may serve as potential signatures related to active UC. The genesis of UC is an extremely complex process during which many genetic and epigenetic modifications of driving genes occur.
Among the hub genes we identified, CXCL10 was highlighted as the highest degree of connectivity genes. CXCL10, as well as CXCL13 were reported a significantly increase in IBD patients (19). TIMP1 is a member of the TIMP gene family and encodes matrix metalloproteinases (MMPs), which are key effectors of tissue-injury-mediated T cell. Both genes are involved in the inflammatory response after tissue damage and repair in IBD. TIMP1 has been detected in rat with Crohn’s-like disease (20). Another study (21) based on knock-out mice showed that TIMP1 deficiency can cause a high expression of immune-related genes. CCL19, as a member of CC cytokines, is significantly expressed in Crohn’s disease (CD), and can lead to chemokine microenvironment normally (22). In light of the results above, we further surmised that these genes may represent as candidate biomarkers for UC.
By using GO annotation, we found that the altered genes displayed in the pathological process of inflammatory reactions, such as leukocyte migration, chronic inflammatory response, regulation of inflammatory response, neutrophil chemotaxis, and leukocyte chemotaxis. Based on the GO terms, the main functional and molecular work might focus on CAMs, cytokine-cytokine receptor interaction, chemokine signaling pathway, adipocytokine signaling pathway, PPAR and TGF-β signaling pathway. Consistent with our results, clinical trials have found that CAMs overexpressed in the colonic mucosa and serum of IBD patients, played a key role in inflammatory response (23,24). Lew et al. (25) found a genetic association with adverse events to anti-tumor necrosis factor treatment in IBD patients, and found that one of the signaling pathways was enriched in cytokine-cytokine receptor interaction. Adipocytokine which is a proinflammatory or anti-inflammatory adipose-derived secretory products, is significantly overexpressed in CD patients (26). PPARγ, a subtype of PPAR, mainly exists in the immune system and adipose tissue and is highly expressed in colon tissue (27). Various evidences have revealed that PPARγ is involved in the pathogenesis of CD (28,29). Moreover, the down expression of TGF-β in UC patient may lead to abnormal anti-inflammatory and negative immunoregulatory effects, thereby increasing the expression of related immune cells and inflammatory cells, followed by the disturbance of intestinal mucosal immune function and persistence of inflammation in UC patients (30). Hence, these pathways are critical in the pathogenesis of UC.
Of note, a panel of miRNAs, such as miR-1231 and miR-92b, may play a vital role in onset of UC. Similarly, miR-1231 and miR-92b are differentially expressed in UC-related CRC (31). Notably, dual oxidase 2 (DUOX2) and trefoil factor 1 (TFF1) were potentially targeted by miR-1231, which suggested that miR-1231 may be a key regulator of DUOX2 and TFF1. DUOX2-inactivating mutations can lead to early onset of IBD (32). Shaoul et al. (33) found that TFF1 was overexpressed in the colonic tissue of children with IBD. CCL11, belongs to CC cytokines and potentially targeted of miR-625, was reported as a potential candidate biomarker in UC and CD (34). By using qRT-PCR, we primarily validated the most significantly up/down-expressed miRNAs (miR-92b and miR-625) and two of their target mRNAs on animal experiment. The results were in line with the microarray analysis. So, we speculated that these miRNAs and their target mRNAs may play an important role in UC development. Additionally, MMP1, a member of MMPs, was targeted by miR-1228. By the implication of KEGG enrichment, it was suggested that MMP1 is closely related to PPAR signaling pathway. We inferred that MMP1 may play vital roles in the development of UC by regulating miR-1228/PPAR signaling pathway, which can provide a new perspective for future studies.
However, the present study comes with some limitations. Firstly, the results were obtained from publicly GEO microarray database and the analysis platforms of three GSE datasets were not uniform. Secondly, the samples were limit which may cause the reliability of our conclusion. Further studies with more samples and unified technological detection platform are needed to confirm our results.
Taken together, our current study used comprehensive bioinformatics analysis to determine the mRNA and miRNA expression between active UC and control. A group of miRNAs and their target genes were identified and several of them were preliminarily confirmed on rodent model, which may serve as potential biomarkers related to UC. In addition, we found several important gene functions and pathways, which may help us understand the molecular mechanisms of UC. However, further experimental and functional studies are warranted to determine the exact role and mechanisms of UC.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81704084, 81673982, and 81603529), the Science and Technology Projects of Jiangsu Provincial Bureau of Traditional Chinese Medicine (YB2017002 and YB2015002), the Natural Science Foundation of the Jiangsu Higher Education Institutions (16KJB360002), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_1541), and sponsored by Qing Lan Project, Chinese medicine advantage discipline funding project and China Scholarship Council (CSC).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Ungaro R, Mehandru S, Allen PB, et al. Ulcerative colitis. Lancet 2017;389:1756-70. [Crossref] [PubMed]
- Wei SC, Chang TA, Chao TH, et al. Management of ulcerative colitis in Taiwan: consensus guideline of the Taiwan Society of Inflammatory Bowel Disease. Intest Res 2017;15:266-84. [Crossref] [PubMed]
- Bopanna S, Ananthakrishnan AN, Kedia S, et al. Risk of colorectal cancer in Asian patients with ulcerative colitis: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2017;2:269-76. [Crossref] [PubMed]
- Thomas S, Hoxha K, Alexander W, et al. Intestinal barrier tightening by a cell-penetrating antibody to Bin1, a candidate target for immunotherapy of ulcerative colitis. J Cell Biochem 2019;120:4225-37. [Crossref] [PubMed]
- Varga ZV, Zvara A, Farago N, et al. MicroRNAs associated with ischemia-reperfusion injury and cardioprotection by ischemic pre- and postconditioning: protectomiRs. Am J Physiol Heart Circ Physiol 2014;307:H216-27. [Crossref] [PubMed]
- Szebeni GJ, Vizler C, Kitajka K, et al. Inflammation and Cancer: Extra- and Intracellular Determinants of Tumor-Associated Macrophages as Tumor Promoters. Mediators Inflamm 2017;2017:9294018.
- Xu XM, Zhang HJ. miRNAs as new molecular insights into inflammatory bowel disease: Crucial regulators in autoimmunity and inflammation. World J Gastroenterol 2016;22:2206-18. [Crossref] [PubMed]
- Van der Goten J, Vanhove W, Lemaire K, et al. Integrated miRNA and mRNA expression profiling in inflamed colon of patients with ulcerative colitis. PLoS One 2014;9:e116117. [Crossref] [PubMed]
- Ţieranu CG, Dobre M, Manuc TE, et al. Gene expression profile of endoscopically active and inactive ulcerative colitis: preliminary data. Rom J Morphol Embryol 2017;58:1301-7. [PubMed]
- Planell N, Masamunt MC, Leal RF, et al. Usefulness of Transcriptional Blood Biomarkers as a Non-invasive Surrogate Marker of Mucosal Healing and Endoscopic Response in Ulcerative Colitis. J Crohns Colitis 2017;11:1335-46. [Crossref] [PubMed]
- Lin X, Li J, Zhao Q, et al. WGCNA Reveals Key Roles of IL8 and MMP-9 in Progression of Involvement Area in Colon of Patients with Ulcerative Colitis. Curr Med Sci 2018;38:252-8. [Crossref] [PubMed]
- Kumagae Y, Hirahashi M, Takizawa K, et al. Overexpression of MTH1 and OGG1 proteins in ulcerative colitis-associated carcinogenesis. Oncol Lett 2018;16:1765-76. [PubMed]
- Ma J, Yin G, Lu Z, et al. Casticin prevents DSS induced ulcerative colitis in mice through inhibitions of NF-kappaB pathway and ROS signaling. Phytother Res 2018;32:1770-83. [Crossref] [PubMed]
- Liu D, Huo X, Gao L, et al. NF-kappaB and Nrf2 pathways contribute to the protective effect of Licochalcone A on dextran sulphate sodium-induced ulcerative colitis in mice. Biomed Pharmacother 2018;102:922-9. [Crossref] [PubMed]
- Cardinale CJ, Wei Z, Li J, et al. Transcriptome profiling of human ulcerative colitis mucosa reveals altered expression of pathways enriched in genetic susceptibility loci. PLoS One 2014;9:e96153. [Crossref] [PubMed]
- Zhao X, Fan J, Zhi F, et al. Mobilization of epithelial mesenchymal transition genes distinguishes active from inactive lesional tissue in patients with ulcerative colitis. Hum Mol Genet 2015;24:4615-24. [Crossref] [PubMed]
- Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Singh UP, Singh NP, Murphy EA, et al. Chemokine and cytokine levels in inflammatory bowel disease patients. Cytokine 2016;77:44-9. [Crossref] [PubMed]
- Talapka P, Berko A, Nagy LI, et al. Structural and molecular features of intestinal strictures in rats with Crohn's-like disease. World J Gastroenterol 2016;22:5154-64. [Crossref] [PubMed]
- Breynaert C, de Bruyn M, Arijs I, et al. Genetic Deletion of Tissue Inhibitor of Metalloproteinase-1/TIMP-1 Alters Inflammation and Attenuates Fibrosis in Dextran Sodium Sulphate-induced Murine Models of Colitis. J Crohns Colitis 2016;10:1336-50. [Crossref] [PubMed]
- Middel P, Raddatz D, Gunawan B, et al. Increased number of mature dendritic cells in Crohn's disease: evidence for a chemokine mediated retention mechanism. Gut 2006;55:220-7. [Crossref] [PubMed]
- Gu P, Theiss A, Han J, et al. Increased Cell Adhesion Molecules, PECAM-1, ICAM-3, or VCAM-1, Predict Increased Risk for Flare in Patients With Quiescent Inflammatory Bowel Disease. J Clin Gastroenterol 2017;51:522-7. [Crossref] [PubMed]
- Arijs I, De Hertogh G, Machiels K, et al. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Am J Gastroenterol 2011;106:748-61. [Crossref] [PubMed]
- Lew D, Yoon SM, Yan X, et al. Genetic associations with adverse events from anti-tumor necrosis factor therapy in inflammatory bowel disease patients. World J Gastroenterol 2017;23:7265-73. [Crossref] [PubMed]
- Paul G, Schaffler A, Neumeier M, et al. Profiling adipocytokine secretion from creeping fat in Crohn's disease. Inflamm Bowel Dis 2006;12:471-7. [Crossref] [PubMed]
- Willson TM, Brown PJ, Sternbach DD, et al. The PPARs: from orphan receptors to drug discovery. J Med Chem 2000;43:527-50. [Crossref] [PubMed]
- Kong R, Luo H, Wang N, et al. Portulaca Extract Attenuates Development of Dextran Sulfate Sodium Induced Colitis in Mice through Activation of PPARgamma. PPAR Res 2018;2018:6079101.
- Yao J, Lu Y, Zhi M, et al. Dietary n3 polyunsaturated fatty acids ameliorate Crohn's disease in rats by modulating the expression of PPARgamma/NFAT. Mol Med Rep 2017;16:8315-22. [Crossref] [PubMed]
- Ihara S, Hirata Y, Koike K. TGF-beta in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J Gastroenterol 2017;52:777-87. [Crossref] [PubMed]
- Tan YG, Zhang YF, Guo CJ, et al. Screening of differentially expressed microRNA in ulcerative colitis related colorectal cancer. Asian Pac J Trop Med 2013;6:972-6. [Crossref] [PubMed]
- Parlato M, Charbit-Henrion F, Hayes P, et al. First Identification of Biallelic Inherited DUOX2 Inactivating Mutations as a Cause of Very Early Onset Inflammatory Bowel Disease. Gastroenterology 2017;153:609-11.e3. [Crossref] [PubMed]
- Shaoul R, Okada Y, Cutz E, et al. Colonic expression of MUC2, MUC5AC, and TFF1 in inflammatory bowel disease in children. J Pediatr Gastroenterol Nutr 2004;38:488-93. [Crossref] [PubMed]
- Adar T, Shteingart S, Ben-Ya'Acov A, et al. The Importance of Intestinal Eotaxin-1 in Inflammatory Bowel Disease: New Insights and Possible Therapeutic Implications. Dig Dis Sci 2016;61:1915-24. [Crossref] [PubMed]